Research Article - Der Pharma Chemica ( 2025) Volume 17, Issue 2
A Study on In-Silico Supported Nephroprotective Action of Coccinia Species
Mayur A Sonar*, Vipul M Patil and Sachin V PatillMayur A Sonar, Department of Pharmaceutical Quality Assurance, Ashokrao Mane College of Pharmacy, Peth-Vadgaon, Tal-Hatkanangle, Kolhapur, India, Email: sonarmayur99@gmail.com
Received: 10-Dec-2023, Manuscript No. DPC-23-122443; Editor assigned: 13-Dec-2023, Pre QC No. DPC-23-122443; Reviewed: 29-Dec-2023, QC No. DPC-23-122443; Revised: 21-Feb-2025, Manuscript No. DPC-23-122443; Published: 28-Feb-2025, DOI: 10.4172/0975-413X.17.2.626-637
Abstract
There are many health problems that can be addressed using medicinal plants. The healthcare system often utilizes herbal remedies, with one particularly important plant being the Coccinia species. This plant has historically been used by various tribes as a treatment for diseases. The species has been shown to possess numerous helpful assets, counting antibacterial, analgesic, antispasmodic, anti-inflammatory, antipyretic, cathartic, and expectorant properties. Chemical components such as alkaloids, glycosides, triterpenoids, phenols, flavonoids, saponins, and tannins can be found in this species. The aim of the present investigation was to isolate, purify, and identify active chemical complex from the leaves of the Coccinia grandis. The extraction process involved consuming Soxhlet apparatus to extract the leaves. Numerous phytochemical tests were conducted, and both TLC and column chromatography were used to isolate the compound. The isolated compounds were further characterized through UV, IR and analysis. The nephroprotective activity of the compound was then evaluated in an in vitro model.
Keywords
Coccinia grandis; Nephroprotective activity; Molecular docking
Introduction
The new year brings worldwide interest in the research and use of traditional medicine. Therefore, governments are trying to cooperate to identify and use effective and efficient medicines in national health systems. Since ancient times, people have studied nature, especially plant, to find new medicines, which has led to the use of traditional medicine, many species of plants effective in treating various diseases. In India, about 95% of traditional medicine is like this. Unani, Ayurveda, Homeopathy and Sadhana are herbs. Plants produce primary and secondary metabolites with different functions. Coccinia is a plant from the Cucurbitaceae family. It is in Central Africa, Asia and India. It is a perennial climbing plant that can be propagated vegetative from seeds. Seeds can be an excellent source of oil and protein that can meet the needs of the market and the consumer. The stalks are either perennially thin climbers or herbaceous climbers, and occasionally, adventitious roots sprout along ground. Branches are long, taut and flexible and can wrap around the entire length of the host. Its leaves are arranged together, its flowers are large, star shaped, white, and its fruit is dark red (Figure 1). The leaves and fruits that were used in foods [1-5].
Coccinia grandis (ivy gourd) is rarely grown as a garden vegetable in tropical and subtropical climates worldwide. Its homeland is thought to be Central Africa, India and Asia. Its long history of human use, cultivation, and transportation obscures its origins. It is a plant species that grows in Southeast Asia. Urolithiasis is the formation of calculi, or condition associated with urinary calculi. The term calculi are synonymous with uroliths, stones, or crystals. These calculi are formed by deposits of polycrystalline aggregates composed of varied amount of crystalloid and organic matrix. Common components of calculi include calcium oxalate (pure), calcium oxalate in combination with as part ate, uric acid (pure), calcium oxalate in combination with uric acid, calcium oxalate dihydrate in combination calcium phosphate. In this present study three plants are selected for the anti urolithiasis study [6-9].
Nephrotoxicity is caused by regular intake of certain drugs. In this case, the kidneys will not be able to clear excess urine and waste products. The kidney is an important organ that helps the body eliminates waste and toxins. Kidney failure is a disease in which harmful metabolites are present in the body and causes serious consequences such as liver enlargement, edema and ascites. The glomerular filtration rate is about 125 ml/minute and the blood flow to the kidney is about 625 ml/minute. As a result, the kidneys receive around four times as much blood as other organs. The glomerular filtration in the nephron's Bowman capsule filters at a rate of around 125 milliliters per minute, therefore the kidney filters approximately 180 L of blood every day [10].
Materials and Method
The whole plant of Coccinia Grandis was collected in the month of Jan 2023 from Bhalwani, Pandharpur, Dist Solapur, India. The plant material was authenticated at Padabhushan Vasantrao Patil Mahavidyalaya, Kavathemahankal, Dist-Sangli [11-15].
Extraction
About 2 kg of shade dried plant leaves of Coccinia grandis extracted in soxhlet successively extracted with n-hexane, chloroform, ethyl acetate and methanol. Each extract was evaporated by using rotary vacuum evaporator. The extract obtained with each solvent was weighed and the percentage yield was calculated in terms of dried weight of the plant leaves. The consistency and colour of the extract were noted. All the solvents used for this work were of analytical grade (Table 1). The n-hexane, chloroform, ethyl acetate, methanol extracts of the leaf powder of Coccinia grandis stamineus were subjected to qualitative chemical analysis [16].
| Sr.no | Extract | Colour | Physical nature | Percentage yield (w/w) |
|---|---|---|---|---|
| 1 | n-Hexane | Green/Sticky mass | Waxy Semisolid | 1.8 |
| 2 | Chloroform | Green/Sticky mass | Semisolid | 2 |
| 3 | Ethyl Acetate | Brownish green solid | Solid | 3.4 |
| 4 | Methanol | Brownish green solid | Solid | 7.9 |
Table 1: The percentage yield of successive extracts of the leaves of Coccinia grandis.
Results
Qualitative chemical examination of plant-based constituents in Coccinia grandis leaf extracts as shown in Table 2 [17].
| Sr.no | Test | n-Hexane | Chloroform | Ethyl acetate | Methanol |
|---|---|---|---|---|---|
| 1 | Alkaloids | - | - | + | - |
| 2 | Carbohydrate | - | - | + | - |
| 3 | Glycosides | - | + | - | + |
| 4 | Phytosterol | - | - | - | + |
| 5 | Fixed oils and Fats | - | - | - | - |
| 6 | Tannins | - | - | - | - |
| 7 | Phenols | - | - | + | |
| 8 | Proteins | - | - | + | - |
| 9 | Gums and Mucilages | - | - | - | - |
| 10 | Flavonoids | - | - | - | + |
| 11 | Terpenoids | - | - | - | + |
| 12 | Steroids | - | + | - | - |
| 13 | Saponins | - | - | + | + |
| Note: +ve indicates positive result; whereas -ve indicates negative result | |||||
Table 2: Phytochemical screening of Coccinia grandis.
Qualitative preliminary phytochemical analysis of Coccinia grandis was performed initially with different chemical reagents to detect the nature of phytoconstituents and their presence in each extract (Table 3 and Figure 2).
Thin-layer chromatography
| S.no | Name of the Extract | Solvent system | No of spots | Rf Values |
|---|---|---|---|---|
| 1 | Methanol (CGM) | Methanol : Ethyl acetate: Water (6:3:1) | 1 | 0.33 |
Table 3: The TLC studies of methanol extracts of Coccinia grandis.
Column chromatography
The Figure 3 show the column chromatography of a) Chloroform b) Methanol c) n-Hexane extract.
- Adsorbent: Silica for column chromatography activated at 110°c for 1 hour
- Length of column: 40 cm
- Length of adsorbent: 25 cm
UV spectrum: Fraction in pet methanol
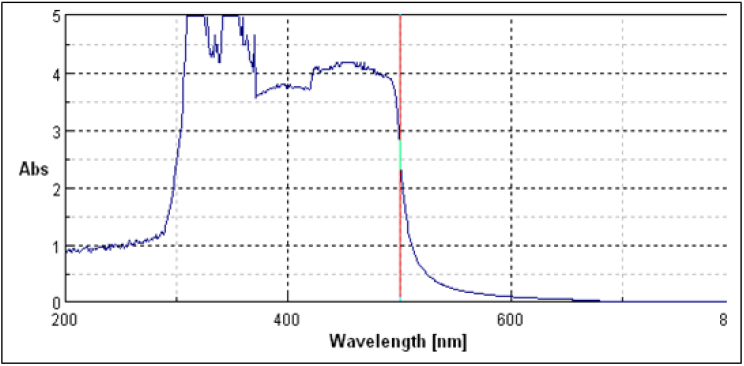
The UV spectrum show in Figure 4.
The UV spectra of isolated cextract from Coccinia grandis was performed using methanol. The UV spectrum showed characteristic bands of fraction in methanol at λ=321,352,413 The UV spectra of standard showed one characteristic peak at λ= 352 (Figure 5 and Table 4).
FTIR of Coccinia grandis extract
| Sr.no. | Peak values | Bonds | Functional groups |
|---|---|---|---|
| 1 | 2952.75 | O–H stretch, H–bonded | Alcohols, Phenols |
| 2 | 2921.35 | C–H stretch | Alkanes |
| 3 | 1493.22 | C–C stretch (in–ring) | Aromatics |
| 4 | 1378.31 | C–O stretch | Alcohols |
| 5 | 842.46, 698.95 | C–H “oop” | Aromatics |
Table 4: FTIR Spectrum analysis.
In-Vitro nephroprotective assay
NRK 52 E Cell line (Normal Kidney Cell Line)
The result show in Table 5 and Figures 6-8.
| Sr.no | Sample code | Conc.(µg/ml) | OD | Mean | % Of inhibition | % Of viability | IC50 (µg/ml) | ||
|---|---|---|---|---|---|---|---|---|---|
| 1 | Control | 1.532 | - | - | - | - | |||
| 2 | Standard | 20 | 1.453 | 1.452 | 1.454 | 1.453 | 5.15% | 94.84% | NE |
| (5,Flurour -acil) | 40 | 1.397 | 1.396 | 1.395 | 1.396 | 8.87% | 91.13% | ||
| 60 | 1.386 | 1.387 | 1.388 | 1.387 | 9.46% | 90.54% | |||
| 80 | 1.369 | 1.368 | 1.369 | 1.368 | 10.70% | 89.30% | |||
| 100 | 1.35 | 1.352 | 1.35 | 1.35 | 11.87% | 88.12% | |||
| 3 | CG1 | 20 | 1.37 | 1.371 | 1.372 | 1.371 | 10.50% | 89.50% | NE |
| 40 | 1.342 | 1.341 | 1.34 | 1.341 | 12.46% | 87.54% | |||
| 60 | 1.287 | 1.286 | 1.285 | 1.286 | 16.05% | 83.95% | |||
| 80 | 1.26 | 1.262 | 1.26 | 1.26 | 17.75% | 82.25% | |||
| 100 | 1.215 | 1.214 | 1.213 | 1.214 | 20.75% | 79.25% | |||
| *NE-Not Evaluable | |||||||||
Table 5: EffectofcompoundagainstNRK52ECellline.
At the different Concentrations Sample Code CG1 shows low percent of inhibition andagainst NRK 52E (Kidney Cells) cell line versus a typical medication 5FU. On basis of percent of viability wecan conclude that the sample CG1 may not harm the normal kidney cell lines and it may protect the kidney from the harm organ damages [18].
In-silico study
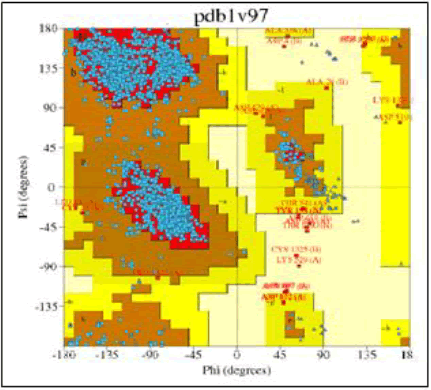
Investigation of protein 1V97 is done by using PDB sum software with the help of Ramchandran plot procheck analysis. According to Ramchandran plot statistics for 1V97 protein the most favoured regions is 88.7%, additional allowed region is 10.3% generally allowed region is 0.6% and disallowed region is 0.4% which are shown in Figures 9 and 10 [19].
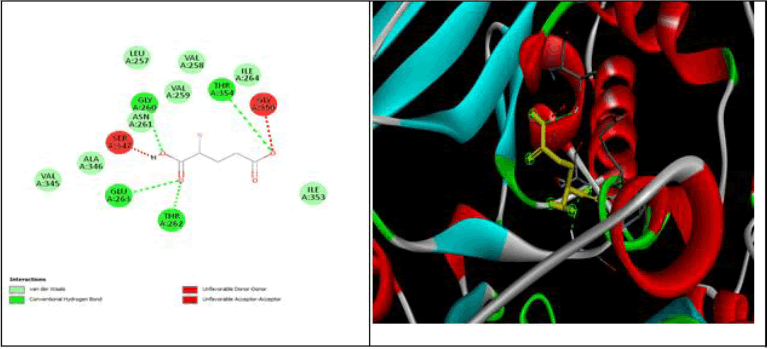
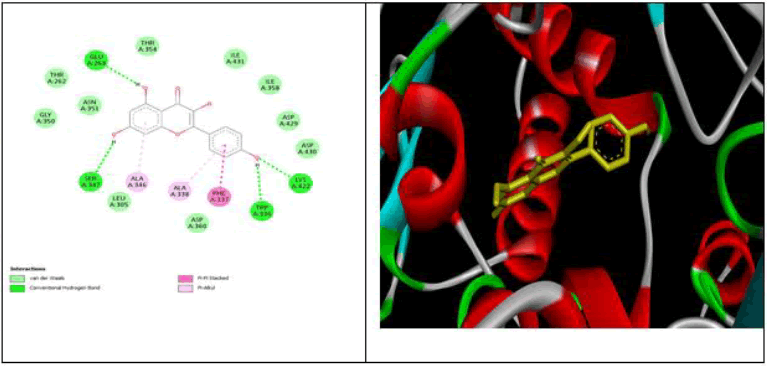
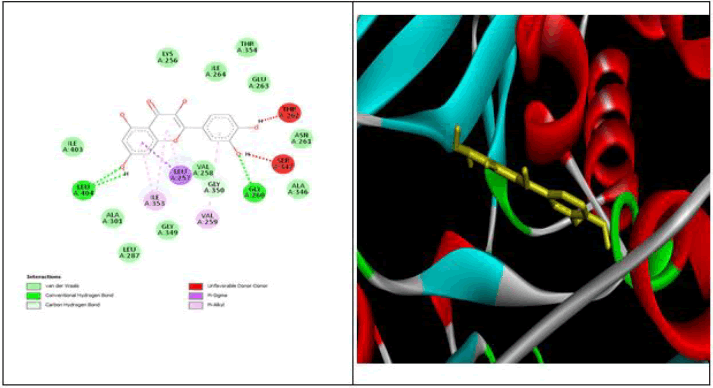
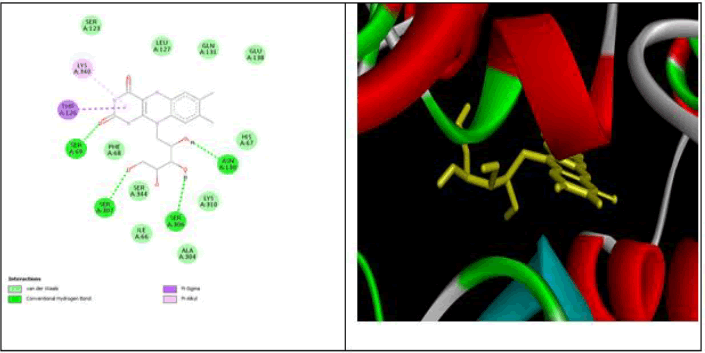
All data obtained from molecular docking of both proteins with ligands are depicted in Tables 6-9 and Figures 11-15 [20].
| S. no. | Compound name | Molecular weight | Log p | H-bond donar | H-bond acceptor | Molar refractivity |
|---|---|---|---|---|---|---|
| 1 | Betasitosterol | 414.71 g/mol | 4.79 | 1 | 1 | 133.23 |
| 2 | Cucurbitacin B | 558.70 g/mol | 1.46 | 3 | 8 | 150.94 |
| 3 | Kaempferol | 286.24 g/mol | 1.47 | 4 | 6 | 76.01 |
| 4 | Thiamine | 265.35 g/mol | -1.6 | 2 | 3 | 73.12 |
| 5 | Ferulic acid | 194.18 g/mol | 1.62 | 2 | 4 | 51.63 |
| 6 | Quercetin | 302.24 g/mol | 1.63 | 5 | 7 | 78.03 |
| 7 | Ombium | 330.29 g/mol | 2.83 | 3 | 7 | 51.63 |
| 8 | Ascorbic acid | 176.12 g/mol | 0.39 | 4 | 6 | 35.12 |
| 9 | Trans pcoumaric acid | 164.16 g/mol | 0.95 | 2 | 3 | 45.13 |
| 10 | Carbonic Acid | 62.02 g/mol | -0.22 | 2 | 3 | 10.65 |
Table 6: Lipinski properties of bioactive compounds from C. grandis leaves.
| S. no | Compound name | Formula | Distribution | Absorption | Toxicity | |||
|---|---|---|---|---|---|---|---|---|
| BBB Permeability | GI Absorption | Human intestinal absorption | Skin permeability | CACO-2 Permeability | Toxicity predicted LD 50 | |||
| 1 | Betasitosterol | C29H50O | 0.781 | Low | 94.464 | -2.783 | 1.201 | 2.552 |
| 2 | Cucurbitacin B | C32H46O8 | -1.003 | Low | 89.52 | -3.496 | 0.588 | 2.381 |
| 3 | kaempferol | C15H10O6 | -0.939 | High | 74.29 | -2.735 | 0.032 | 2.449 |
| 4 | Thiamine | C12H17N4O5 | -0.368 | High | 100 | -2.792 | 0.867 | 2.672 |
| 5 | Ferulic acid | C10H10O4 | -0.239 | High | 93.685 | -2.72 | 0.176 | 2.282 |
| 6 | Quercetin | C15H10O7 | -1.098 | High | 77.207 | -2.735 | -0.229 | 2.471 |
| 7 | Ombiun | C17H14O7 | -1.089 | High | 87.47 | -2.735 | 0.402 | 2.272 |
| 8 | Ascorbic acid | C5H8O6 | -0.985 | High | 39.154 | -2.955 | -0.255 | 1.063 |
| 9 | trans p coumaric acid | C9H8O3 | -0.225 | High | 93.494 | -2.715 | 1.21 | 2.155 |
| 10 | Carbonic ssacid | CH2O3 | -0.428 | High | 83.064 | -2.737 | 1.12 | 1.439 |
| 11 | Niacin | C6H2ON4O6 | -0.323 | High | 94.099 | -2.735 | 1.17 | 2.24 |
Table 7: ADME properties of various bioactive compounds.
| Compound Name | Binding affinity (kcl/mol) | Conventional H- bond Interaction | Pi Sigma Interaction | Pi Alkyl interaction | Van-der-Waals interaction | Other interaction |
|---|---|---|---|---|---|---|
| Betasitosterol | -9.3 | GLU 411 | - | VAL518,LEU81,LEU140 , LEU139 | GLU143,TYR69, ASN70, ASN 66, VAL351,ASN136, ASN85 ARG124, SER516, TRP357, SER355, TYR523 | - |
| Cucurbitacin B | -10 | ILE 204, ASN211, GLU124 | - | - | ALA 207, TRP220, TYR135, LEU139, SER219, ALA208 | - |
| Kaempferol | -8.2 | ASP415, TYR520, LYS511, GLN281, ASP377 | VAL380 | ALA 354 | GLN369, GLU162, HIS353, HIS513, PHE457, TYR523, PHE527, HIS383 | - |
| Thiamine | -6.9 | TYR 62, SER 355 | - | - LEU 82, LEU 139,LEU 81, LEU 140 | TYR 69, VAL 351, TRP 357 ,GLU 143 ,ASN 70, PHE 512, VAL 518 ,ASN 85,ASN 136 | Other C- H bond-LYS 368, SER 516 |
| Ferulic Acid | -6.5 | ARG 124, SER 517 | - | ILE 204, ALA 207 | LEU 139, TYR 135, SER 219, TYR 213,ASP 121,ALA 208 | Other Pi-anion- GlU123 Amide-pistacked-TRP 220 |
| Quercetin | -8.3 | THR 301, ASP 453 | - | LYS 449, LEU 375 | THR 302, ASP 300, MET 299, LEU 433, SER 298 ,GIU 376, VAL 379, MET 450,SER 284, ASN 285 | - |
| Ombiun | -8.5 | GLY 404,GLU 384. TYR 523, ARG 522 | - | MET 223, PRO 407, PHE 512, HIS 353, VAL 518 | ASN 406, PHE 391, ALA 356, SER 355 | Other Pi-cation-GLU 411 Pi-Anion-GLU 403C-H bond HIS 410, HIS 387, HIS 513 |
| Ascorbic acid | -5.7 | ASP 121, TRP 220 | - | - | TYR 213, ALA 208, ALA 207, ILE 204,ARG 124, TYR 135 ,GLU 123,ASN 211, SER 219 | |
| trans p coumaric acid | -6.2 | ARG 124, SER 517 | - | ALA 207 | LEU 139, TYR 135,SER 219,ASP 121 ,TYR 213, ASN 211, TRP 220, ILE 204 | Other Pi-anion- GLU 123 |
| Carbonic Acid | -3.3 | HIS 353, CYS 352, SER 147, VAL 350 | - | - | PHE 512, VAL 351, LEU 161, TYR 146 | - |
| Niacin | -5.4 | ASN 85, ARG 124 | - | - LEU 139 | LEU 81, TYR 62, ASN 136 ,LEU 140, GLU 143 | - |
| Quercetin | -8.3 | THR 301, ASP 453 | - | LYS 449, LEU 375 | THR 302, ASP 300, MET 299, LEU 433, SER 298 ,GIU 376, VAL 379, MET 450,SER 284, ASN 285 | - |
Table 8: Moleculardocknig result examination of bioactive substances of coccinia grandis against the protein 1UZE.
| Sr. no. | Compound | Lipinski | Ghose | Veber | Egan | Muegge |
|---|---|---|---|---|---|---|
| 1 | Betasitosterol | Yes | No | Yes | No | No |
| 2 | Cucurbitacin B | Yes | No | Yes | No | Yes |
| 3 | Kaempferol | Yes | Yes | Yes | Yes | Yes |
| 4 | Thiamine | Yes | Yes | Yes | Yes | Yes |
| 5 | Ferulic Acid | Yes | Yes | Yes | Yes | No |
| 6 | Quercetin | Yes | Yes | Yes | Yes | Yes |
| 7 | Ombium | Yes | Yes | Yes | Yes | Yes |
| 8 | Ascorbic acid | Yes | No | Yes | Yes | No |
| 9 | Trans p coumaric acid | Yes | Yes | Yes | Yes | No |
| 10 | Carbonic Acid | Yes | No | Yes | Yes | No |
| 11 | Niacin | Yes | No | Yes | Yes | No |
Table 9: Drug likeliness propertiesof isolated ligand from coccinia grandi.
Conclusion
Several polarity phytoconstituents were extracted using a soxhlet device and solvents such as nhexane, chloroform, ethyl acetate, and methanol in order to perform a phytochemical analysis on the leaf powder of Coccinea grandees. An occurrence in Saponins, phenols, flavonoids, terpenoids, glycosides, and phytosterol was demonstrated by the methanolic extract. The UV spectra of isolated extract from Coccinia grandiswas performed using methanol. The UV spectrum showed characteristic bands of fraction in methanol at λ=321,352,413The UV spectra of standard showed one characteristic peak at λ=352. The FTIR analysis of Coccinia grandis gave broad peak at 2952.75cm-1 which indicated the presence of phenolic O-H stretching. It showed strong peaks at2921.35cm-1 which indicated the presence of alkanes whereas the peaks at1493.22 and 1378.31cm-1 attributed to existence of the aromatic ring's C–C stretching and the C–O stretch, respectively. In-vitro antinephroprotective like different Concentrations Sample Code CG1 shows low percent of inhibition and against NRK 52E (Kidney Cells) cell line versus a typical medication 5FU. On basis of percent of viability wecan conclude that the sample CG1 may not harm the normal kidney cell lines and it may protect the kidney from the harm organ damages. In-silico analysis for chemical components of leaf extracts of C. grandis was done Using molecular docking, it was revealed how important drug design is for creating new medications that block targets. Most of biomolecules from C. grandisleaves were shows better docking results against the active sites of the selected receptor 1V97. Binding energy, hydrogen bond interactions and Vander Waal’s interactions for the enzymes are listed in Tables. As the binding energy lowers, binding efficiency will increase. Similarly, as the number of as the number of hydrogen bonds between the ligand and enzyme rises, so does the binding's strength.
References
- Satyavati GV, Gupta AK, Tandon N. Medicinal Plants of India. 1987; 2:304-306.
- Hossain SA, Uddin SN, Salim MA, et al. JSIR. 2014; 3(1): p.65-71.
- Suresh Vc, Narayana Ra, Msc Bose Ag, et al. IJAPSR. 2015; 6 (2): p.2784-2789.
- Sumaryono W, Proksch P, Wray V, et al. Planta Med. 1991; 57(02): p.176-80.
[Crossref] [Google Scholar] [PubMed]
- Skoog Douglas A, James HF, Nieman Timothy A. London, Thomson Brook, 2005; pp. 350-432.
- Willard HH, Merritt LL, Dean JA. New York: D. Van Nostrand company (cannada ) Ltd, 4th ed,1965; pp. 180-445.
- Mezei M. J Mol Graph Model. 2003; 21(5):463-472.
[Crossref] [Google Scholar] [PubMed]
- Febrina EL, Alamhari RK, Asnawi AI, et al. Int J App Pharm. 2021; 13(4):210-215.
- Mathews MM, Sunny B. Int J Pharm Sci Rev Res. 2019; 54(2):29-36.
- Daneman R, Prat A. Cold Spring Harb Perspect Biol. 2015; 7(1): p.a020412.
[Crossref] [Google Scholar] [PubMed]
- Elmeliegy M, Vourvahis M, Guo C, et al. Clin Pharmacokinet. 2020; 59: p.699-714.
[Crossref] [Google Scholar] [PubMed]
- Finch A, Pillans P. Aust Prescr. 2014;37(4): p.137â??139.
- Cheng F, Li W, Zhou Y, et al. J Chem Inf Model. 2012; 52(11): p.3099â??3105.
[Crossref] [Google Scholar] [PubMed]
- Rani VP, Moorthy BP, Priya KS, et al. AJPCT. 2016; 4(4): p.98-105.
- Narayanan M, Jayashree T, Kandasamy S, et al. Process Biochem. 2020; 109: p.178-185.
- Bindurani LR, Singh A. JDDT. 2019; 9(3): p.238-245.
- Asif M, Tariq M, Khan A, et al. J Agric Sci. 2017; 12: p.108-122.
- Jha R, Ramani PT, Patel D, et al. J Med Plants Stud. 2016; 4(3): p.18-22.
- Nalvolthula R, Merugu R, Rudra MP. Int J Chem Tech Res. 2014; 7:2374-2380.
- Mamillapalli V, Khantamneni PL, Mohammad Z, et al. Int J Pharm Sci Res. 2016; 7(10): p.4074-4084.