Research Article - Der Pharma Chemica ( 2019) Volume 11, Issue 2
Docking studies of chalcone analogue compounds as inhibitors for breast cancer MCF7 cell line
Neni Frimayanti*, Benni Iskandar and Regina Dewi PutriNeni Frimayanti, Sekolah Tinggi Ilmu Farmasi Riau, Jalan Kamboja, Simpang Baru Panam, 28292, Indonesia,
Abstract
Molecular docking studies of chalcone analogue with the protein target from the crystal structure modeling of H-RAS P21 Triphospate enzyme (PDB ID: 5P21) was carried out using computational approach with Autodock Vina program. The aims of this study is to determine the activity of 15 chalcone analogue, these compounds were obtained from the previous studies as breast cancer inhibitors and also to compare the activity of several chalcone analogue with the doxorubicin. Doxorubicin is used as a positive control. Based on the docking results, it is showed that compound 4 has a greater potential to be used as an inhibitor of MCF-7 breast cancer cells than the other test compounds. Compound 4 has the binding affinity energy of -9.0 kcal/mol, it is indicated that ligand perform a bonding with the receptor through the eleven active sites of doxorubicin. This compound can be further analyzed especially in vivo test to prove its inhibitory activity.
Keywords
Breast Cancer, Chalcone, MCF-7 cell line, Moleculer docking.
Introduction
Cancer is one of the leading causes of death in the worldwide. Breast cancer is the second most common cancer in the world and this cancer that occurs in women with an estimated 1.67 million (25%) new cancer cases diagnosed in 2012 [1]. In Indonesia, breast cancer is the number two cancer after cervical cancer and there is a tendency from year to year in the incidence increases. The number of breast cancers in Indonesia is approximately 23140 new cases every year [2].
Most likely the caused of breast are related to changes in the genetic material (DNA) in our cells. DNA changes are often related to our lifestyle, but some can be due to age and other factors [3]. The etiology of breast cancer can be explained as variety of interlated factors, such as hormones, the environmental sociobiology and physicology among others can influence breast cancer development [4]. The development of breast cancer is started in any part of breast such as in invasive or infiltrating ductal carcinoma (IDC) and invasive or infiltrating lobular carcinoma (ILC) [5].
There are some medication of cancer such as chemotherapy, radiotherapy and surgery. Chemotherapy is a potential choice for cancer treatment, currently, a cancer chemotherapy agent that is still widely used is doxorubicin. Doxorubicin is included in anthracycline antibiotics. However, the use of doxorubicin can cause side effects including cardiotoxicity which can end with heart failure [6], hepatotoxicity [7] and resistance [8].
MCF-7 is a breast cancer cell taken from the site of breast cancer pleural metastatic effusion in a 69-year-old Caucasian woman [9]. MCF-7 cells are cells that are commonly used to test the effect of breast cancer in vitro because of the best form of all types of human breast cancer cells [10].
Chalcone is a common structure that is widely found in flavonoids [11]. Chalcone or 1,3-diphenyl-2-propene-1-on, in the form of two aromatic rings that are mutually bound [12]. Chalcone is a bioactive compound that has various biological activities such as antibacterials [13], antitumor [14-16], anticancer [17,18] and antioxidants [14]. One new method used to predict the activity of compounds for anticancer is the molecular docking. Molecular docking is a computational procedure that can predict non-covalent bonds of macromolecules (receptors) with a small molecule (ligand), the efficiently using its structure through molecular docking simulations is also can measured [19].
Research on molecular docking was conducted to determine the activity of 15 chalcone analogue compounds which are obtained from Mai et al. [20] with the IC50 value as an inhibitor of breast cancer. Interaction between amino acid residues on receptors and the ligand (i.e. 15 chalcones) is very important to know the reason that compound is active or not. The aim of this study is to determine the activity of 15 chalcone analogue compounds with doxorubicin was used as positive control. The best activity of the chalcone analogues compound was then can be used as new inhibitors for breast cancer and also it can be applied as a reference for alternative medicine.
Materials and Methods
Tools and material
A set of Aspire V5-471G 1.40 GHz computers, with 4096 MB RAM. The program used are Chemdraw Ultra 13.0 (CambridgeSoft), Discovery Studio Visualizer (DSV), MGL Tools 1.5.6 (including AutoDock Tools 1.5.6, AutoDock Vina), and pyMOL.
The materials used for molecular docking are target proteins obtained from the modeling of the crystal structure of the H-RAS P21 Triphospate enzyme in the format of PDB (Protein Data Bank) with 5P21 code with a resolution of 1.35 Å, doxorubicin as positive control and the ligand used is structure of chalcone analogue compounds are presented in Table 1.
| Molecular structure | Molecular structure |
|---|---|
(1)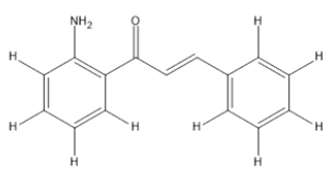 |
(2)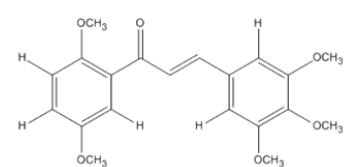 |
(3)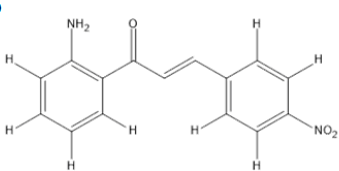 |
(4)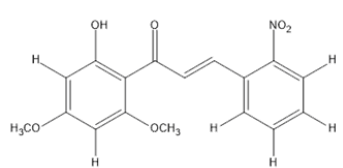 |
(5)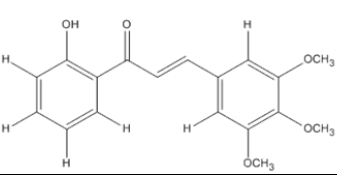 |
(6)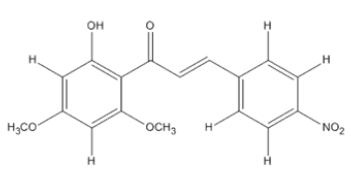 |
(7)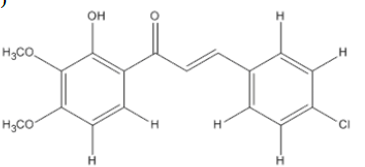 |
(8)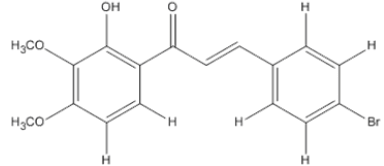 |
(9)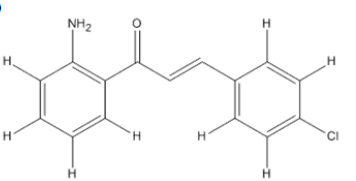 |
(10)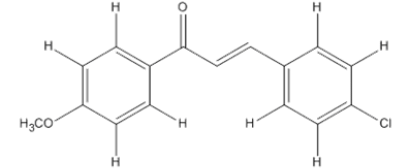 |
Table 1: Molecular ligand structure
Protein preparation
The structure of crystallographic protein or enzyme with PDB-ID 5P21 is downloaded from the site www.pdb.org in PDB format, the water molecules were then removed using discovery studio visualizer (DSV). Preparation is done by adding the hydrogen using AutoDock Tools 1.5.6 and it was saved into PDBQT format.
Ligand preparation
The molecular structure of the chalcone analogues were skeched using ChemDraw Ultra 13.0, then saved in PDB format using DSV. Ligand in PDB format are then inputted to the AutoDock Tools 1.5.6 program for further preparation. Rotating bonds can be corrected in the torsion tree panel for ligand flexible properties. Then the ligand is saved in the form of PDBQT.
Grid box settings
In AutoDock Tools 1.5.6, the Grid menu is selected to open enzymes that have been prepared in the PDBQT format, then the grid box is created to determine the space for docking simulation will be running. The distance size in the grid box is set to 1 Å and the dimensions of the x, y, and z are arranged in such a way that the grid box can load the active site of the enzyme. The data grid box must be kept in mind because it will be needed for creating the configuration data.
Docking simulation using autodock vina
Docking simulation was run using AutoDock Vina. It has been done by entering the commands in the command prompt (cmd) base on the installation location of the AutoDock Vina program and also the work folder on the computer. After completed the running process, in the work folder there will be two files, they are "log.txt” and "out.pdbqt". "log.txt” is the docking results, it is consisted of the affinity value and "out.pdbqt", it is contained the conformation of ligand after docking. The conformation can be separated by entering the split to cmd command to perform one by one of the conformation.
Visualization of docking results with PyMOL and DSV
PyMOL is used to combine the enzymes with ligands from docking into a complex and saved it in PDB format. The interaction between enzymes and ligands can be displayed using the DSV program in 2D and 3D dimension.
Data analysis
The obtained data from the docking process of chalcone analogue compounds will come out the binding free energy with nine ligands conformations. The best pose will selected base on the smallest value of the binding free energy. The interaction between the active site of the protein with the ligands are performed in 2D and 3D using DSV.
Conclusion
Based on docking results, it is showed that compound 4 has a greater potential and can be used as an inhibitor of MCF-7 breast cancer cells. Compound 4 has low energy (affinity) of -9.0 kcal / mol and also this ligand has a stable bond with the receptor and it also has eleven interaction with amino acid like interaction of doxorubicin as a positive control. The important amino acids involved are Leu120, Phe28, Lys117, Ala146, Asp119, Ser145, Gly15, Lys147, Ser17, Pro34, and Tyr32, with 5 types of bonds include van der Waals bonds, hydrogen bonds, carbon-hydrogen bonds, alkyl bonds, and pi-alkyl bonds.
References
- Anonim, Kanker Payudara, Jakarta, Pusat Data dan Informasi Kementerian Kesehatan Republik Indonesia, 2016.
- Suyatno, E.T. Pasaribu, Bedah Onkologi Diagnosis dan Terapi, Jakarta, Penerbit Sagung Seto, 2010.
- G. Xianlong, W. Xiaomin, Y. Jiajie, Z. Jingqun, Z. Weiwei, D.M. Dominic, Z. Yongguo, B. Mamdouh, [21] M. Gregory, S. Jun, Li. Zhenyu, F. Michael, A. Xun, Cardiovas. Res., 2018, 114, 737-746.
- L.T. Angahar, MOJ. Biol. Med., 2017, 1(4), 92-96.
- K. Sun, S. Tang, Y. Hou, H. Yixuan, X. Lei, C. Yanlin, Y. Jiali, P. Meixi, Z. Maojia, C. Xiaojiang, L. Manran, EbioMed., 2019.
- Anonim, Farmakologi dan Terapi, Jakarta, Fakultas Kedokteran Universitas Indonesia, 2012.
- H.I. El-Sayyad, M.F. Ismail, F.M. Shalaby, R.F. Abou-El-Magd, R.L. Gaur, A. Fernando, M.H.G. Raj, A. Ouhtit, Int. J. Biol. Sci.,2009, 5(5), 466-473.
- M. Han, Q. Lv, X.J. Tang, Y.L. Hu, D.H. Xu, F.Z. Li, W.Q. Liang, J.Q. Gao, Journal of Controlled Release.,2012, 163(2), 136-144.
- Anonim, MCF-7 (ATCC® HTB-22™), Breast Carcinoma Cell Line, 2016.
- L. Widowati, H. Mudahar, Media Litbang Kesehatan.,2009, 119(1), 9-14.
- H.O. Saxena, U. Faridi, J.K. Kumar, S. Luqman, M.P. Darikar, K. Shanker, C.S. Chanotiya, M.M. Gupta, A.S. Negi, Elsevier., 2007, 72, 892-900.
- C.B. Patil, S.K. Mahajan, S.A. Katti, J. Pharmaceut. Sci. Res.,2009,1(3), 11-22.
- D. Hariyanti, H.Y. Teruna, Jasril, Jurnal Online Mahasiswa, FMIPA.,2014, 1(2), 352-358.
- B.P. Bandgar, S.S. Jalde, L.K. Adsul, S.N. Shringare, S.V. Lonikar, R.N. Gacche, N.A. Dhole, S.H. Nile, A.L. Shirfule, Med. Chem. Res.,2012, 21, 4512-4522.
- E-H. Zhang, W. Ru-Feng, S-Z. Guo, B. Liu, Hindawi Publishing Corporation,2013.
- A. Abbas, M.M. Naseer, A. Hasan, T.B. Hadda, J. Mater. Environ. Sci., 2014, 5(1), 281-292.
- A. Shah, A.M. Khan, R. Qureshi, F.L. Ansari, M.F. Nazar, S.S. Shah, Int.J. Mol. Sci., 2008, 9, 1424-1434.
- R. Arianingrum, I. Sulistyo, S. Atun, Jurnal Penelitian Saintek., 2011, 16(2), 121-132.
- O. Trott, A.J. Olson, Comput.Chem.,2009, 31(2), 455-461.
- W. Mai Chun, M. Yaeghoobi, N. Abd-Rahman, Y.B. Kang, M.R. Pichika, Eur. J. Med. Chem., 2014, 77, 378-387.
- M. Yaeghoobi, N. Frimayanti, C.F. Cheel, K.K. Ikram, B.O. Najjar, S.M. Zain, Z. Abdullah, H.A. Wahab, N.A. Rahman, Med. Chem. Res.,2016, 25(10), 2133-2142.
- F. Suhud, Siswandono, T. Budiati, Media Pharmaceutica Indonesiana., 2017, 1(3), 154-163.
- F. Suhud, Jurnal Farmasi Indonesia., 2015, 7(4), 242-251.
- N. Frimayanti, B. Iskandar, M. Yaeghoobi, H.C. Han, R. Yusof, S.M. Zain, N.A. Rahman, Asian Pasific J. Trop. Dis., 2017, 7(12), 792-796.