Research Article - Der Pharma Chemica ( 2022) Volume 14, Issue 8
In Silico Evaluation of Selected Phytochemicals for Anti-nCovid Potential Based on Molecular Docking Studies and Their Pharmacokinetics and Drug-likeness Predictions
Selvapraba S*, Venkatachalam T and Senthil Kumar NSelvapraba S, Department of Pharmaceutical Chemistry, JKKMMRF’s –Annai JKK Sampoorani Ammal College of Pharmacy, Komarapalayam, Namakkal –638 183, Tamil Nadu, India, Email: sspraba107@gmail.com
Received: 28-May-2022, Manuscript No. dpc-22-65278; Editor assigned: 01-Jun-2022, Pre QC No. dpc-22-65278; Reviewed: 22-Jun-2022, QC No. dpc-22-65278; Revised: 14-Jul-2022, Manuscript No. dpc-22-65278; Published: 01-Aug-2022, DOI: 10.4172/0975-413X.14.8.36-47
Abstract
SARS-CoV-2 has devastated the world with its rapid spread and fatality. Pharmaceutical giants respective of their market has stretched towards research and enhancing the productive capacities of their units to meet the unprecedented drug demand. Drug discovery approaches involving all in silico, in vitro, in vivo approaches to design and develop has fastened. The present work explains in-silico drug discovery methods like molecular docking and molecular dynamic simulations to screen for highly probable, safe, and effective phytochemical principles against SARS-CoV-2. Docking of various phytochemical principles against three X-ray crystallographic and one electron microscopic structures of SARS-CoV-2 proteins that include the RNA dependent RNA polymerase and 3CL protease (3CL pro) by using the Glide Schrodinger docking software 2019_4. 3.1 was carried out. The best fit drug candidates among the docked ligand structures by their docking score and interactions were selected and subjected to prediction of drug likeliness and ADME parameters. It is observed that the phytochemicals/bioactives such as Scutellarein, Saikosaponin D, Syringaresinol and 5,7,2’,3-Tetramethoxyflavone hold promise in inhibiting the SARS-CoV-2 key viral proteins and displayed the capability to suppress SARS-CoV-2 proteins and justify their further in vitro and in vivo studies. The present study could be the starting point for the future ligands from natural sources in 2019-nCoV RdRp and 3CL pro.
Keywords
Molecular Docking; SARS-CoV-2; Phytoconstituents; Drug likeness; ADME parameters; Scutellarein; Saikosaponin D; Remdesivir; Anti-nCovid; Binding energy; Binding Score; In silico
INTRODUCTION
The mankind was terribly wobbled, troubled and lead to catastrophy in every domain by the most infectious and deadly COVID -19 virus. The entire world has to face a crisis that it may wipe out the human kind. Almost every nuke and corner has thoroughly put under intense pressure to carry out research and validate the findings to contain the infection, be it may be developing sanitizers, PPEs, diagnostic kits, biomedical equipment, and most importantly to design novel agents as well testing the existing ones for repurpose to treat this novel virus. In the coronaviridae family of viruses 2019-nCoV is a novel strain not identified earlier in humans [1]. The initial outburst of 2019-nCoV in the epicenter of Wuhan province in China has spread briskly and affected other parts of China. Soon or a little later the entire world was afflicted and faced enormous challenge and health care burden [2]. The lethality of 2019-nCoV epidemic is unprecedently larger than the severe acute respiratory syndrome (SARS) epidemic. World Health Organization (WHO) has declared nCoV as pandemic on 11 March 2020, and gave the nomenclature as nCovid-19 COVID-19 and declared as ‘Public Health Emergency of International Concern’ (PHEIC), affecting around 212 countries and accounting for nearly 1 million deaths across the globe. In this scenario, WHO has issued the COVID-19 advisory from time to time through WHO website (WHO, 2020). Pharmaceutical giants respective of their market has stretched towards research and enhancing the productive capacities of their units to meet the unprecedented drug demand. Biotech firms and Vaccine manufacturers all over the globe are thoroughly designing and evaluating the vaccines which are a time taking and under stringent regulatory standards. Drug discovery approaches involving all in silico, in vitro, in vivo approaches to design and develop has fastened. In silico paradigms have much larger role in enhancing high throughput screening efforts. Particularly, in-silico studies have great role of finding magic molecules and repurposing of existing drugs in the treatment of COVID-19 [3]. Earlier in 2003 SARS epidemic, the effectiveness of herbal treatments was demonstrated and gained huge prominence. Hence as demanded by the need and thrust, to find the therapeutics to lessen the COVID induced mortality. Complementary and alternative medicine systems were largely focused their research to evolve prospective molecules.
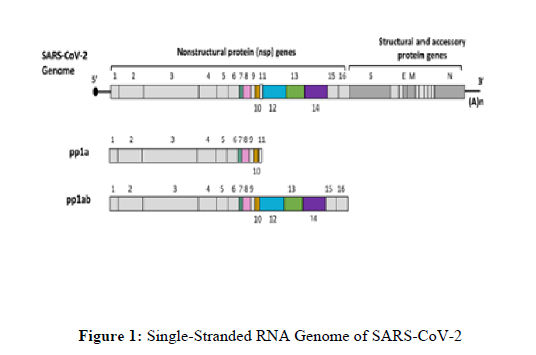
The single-stranded RNA genome of SARS-COV-2 is depicted in Figure 1.
Two-thirds of the genome encodes two large polyproteins, pp1a and pp1ab, that are cleaved into 16 non-structural proteins. The last one-third of the genome encodes structural and accessory proteins. This figure was created with BioRender.
The studies on SARS-CoV-2 as well as previous SARS-CoV and other coronaviruses have mostly identified with the functions of these structural proteins, non-structural proteins as well as accessory proteins, which are summarized in Table 1. Thus, keeping the above view into consideration, the present investigation was undertaken to determine the efficacy of 48 different phytoconstituents in comparison to reference drugs (Remdesivir) against two different protein targets that is, RNA dependent RNA polymerase and 3CL pro using in-silico docking.
Table 1: Coronaviruses Proteins and their functions
| Proteins | Function | Experiment PDB ID |
|---|---|---|
| NSP1 (Leader protein) | NSP1 is 180 amino-acids (aa) containing polypeptide inhibits host gene translation by interacting with 40s ribosomal subunit. Suppresses host gene expression and facilitates viral gene expression in infected cells [4]. | 7k3n |
| NSP2 | NSP2 contains 638-residue polypeptides Interacts with the host factors like prohibitin1 and prohibitin2, thereby disrupt the functional integrity of host mitochondria. NSP-2 also disturbs host intracellular environment and signalling [5]. |
Prediction [6] |
| NSP3(Papain like protease) | NSP3 is a polypeptide with 1945residues that include papain-like protease (PLpro) and multi-pass membrane protein. PLpro helps for cleaving and releasing NSP1, NSP2, and NSP3. PLpro participates in deubiquitination reaction and evades host cell response. NSP-3 along with NSP-4 promotes the formation of virally induced cytoplasmic double-membrane vesicles, necessary for viral replication. NSP3 also antagonises generation of type I interferons [7]. |
Partially available from experiments: Residues 1570- 1877 (PLpro, PDB ID: 7kol, etc.); Residues 819-929 (PDB ID: 7kag, etc.); Residues 1024- 1192 (PDB ID: 6wcf [8], etc.) |
| NSP4 | NSP4 is a polypeptide multi pass membrane with 500 amino-acids. NSP3 promotes formation of virally induced cytoplasmic double membrane vesicle for RNA replication [9]. |
Prediction [10] |
| NSP5(3c- like proteinase) | NSP5 is a 306-residue main protease (3CLpro) of SARS-COV-2. It cleaves and releases NSP4-NSP16. It identifies core sequence apart from binding ADP-ribose-1"-phosphate (ADRP) and in the maturation of NSP [11]. |
Experiment (PDB ID: 5r84 [12], etc.) |
| NSP6 | NSP6 is a 290-residue component which limits the expansion of autophagosomes that are no longer delivered to lysosomes [13,14]. | Prediction [10] |
| NSP7 | NSP7 is a 83-residue peptide along with NSP8 participates in viral replication by serving as primase [15]. | 6m5i,etc.) |
| NSP8 | NSP8 is a 198-residue component that together with NSP7 helps in viral RNA synthesis [16]. | 6m5i, etc.) |
| NSP9 | NSP9 is a 113-residue component serves as a dimeric ssRNA binding protein [16]. | 6w4b, etc.) |
| NSP10 | NSP10 is a 139-residue plays a pivotal role in viral transcription by interacting with NSP14 and NSP16 to stimulate the activities of 3'-5' exoribonuclease and 2'-O-methyltransferase in viral mRNA cap methylation [16]. | 6zct [17], etc.) |
| NSP11 | NSP11 is a 13-residue component that helps in the formation ofN-terminal of NSP-12 [16]. | NA |
| NSP12 (RNA-dependent RNA polymerase) | It is a 932-residue which is RNA-dependent RNA polymerase (RdRp) that performs dual function of replication and transcription of viral genome [18]. Helps in synthesis of complementary RNA strand. Currently FDA approves drugs inhibiting RdRp like Remdesivir and Flavipiravir [17]. |
6m71 , etc.) |
| NSP13 (Helicase) | A multifunctional protein helping in viral genome replication along with NSP12. It is associated with viral nucleoprotein [19]. |
5rlh [18], etc.) |
| NSP14 | NSP14 is a 527 residue by interacting with NSP10 helps for viral transcription by processing exoribonuclease activity and (guanine-N7) methyl transferase (N7-MTase) activity [20]. | Prediction [6] |
| NSP15 (endoRNAse) | NSP15 is a 346-residue polypeptide helps for evasion of virus by host cell dsRNA sensors [21]. | 5s72, etc.) |
| NSP16 | NSP16 is a 298-residue polypeptide together with NSP10participates in mRNA cap 2’-O-ribose methylation to the 5’-cap structure [23]. | 6w4h, etc.) |
| Spike protein (Surface glycoprotein) | Spike protein having 1273 amino-acids aids to down regulate host tetherin by lysosomal engulfment leading to blocking its antiviral activity. Subunit S1 holds the virion to the host cell membrane through angiotensin-converting enzyme 2 (ACE2) and internalization of the virus into the endosomes of the host cell. The stalk region of S is three hinged having unexpected freedom to orient into space. It utilizes human TMPRSS2 for priming with human lung cells. S2 helps in fusion of the virion and host cell membrane thus acting as fusion protein. The S protein exists in three conformational states: native state, pre-hairpin intermediate state, and post-fusion hairpin state. This arrangement places the fusion peptide in close proximity with the c-terminal of ectodomain leading to fusion of viral and hosts cell membrane [24]. |
7c2l [24], etc.) |
| ORF3a | It is a multi-pass membrane protein having 275 residues that forms potassium sensitive ion channel known as viroporin. This ORF3a activates expression of FGA, FGB and FGG in lung epithelium followed by induction of apoptosis and downregulation type-1 interferon. It also aids for the ubiquitination of IFN alpha-receptor subunit 1 (IFNAR1) thereby stimulation of NF-kB and NLRP3 pathways inducing cytokine storm which is fatal for humans. Apart from thisORF3a contributes in virion release [25]. | 6xdc [26] |
| ORF3b | ORF3b is a 22-amino-acid protein that, along with nucleocapsid protein may inhibit the induction of IFN-1. This residue was found in SARS-CoV-2 related genomes in pangolins and bats [27]. | NA |
| ORF4 (Envelope protein) | ORF4 is an envelope protein with 75 amino acid residues playing a dominant role in viral assembly and maturation. This envelope protein forms an ion transport channel by interacting with host membrane. This protein is known to cause apoptosis [28]. | Partially available from experiment: Residues 8-38 (PDB ID:7k3g [29]) |
| ORF5 (Membrane protein) | ORF5 is a 222-residue membrane protein highly conserved. It shares some functions of ORF4 by involving with ER-Golgi apparatus of host cell. The packaging of viral genome into virion is also carried out by this protein [30]. | Prediction [10] |
| ORF6 | ORF6 is a virulence factor of61 residues. It disrupts cell nuclear import complex by interacting karyopherin alpha 2 and karyopherin beta 1. Hence the import a factor doesn’t translocates to nucleus prevents STAT pathway that generally activates due to production of interferons enabling the virus survival and host cell to apoptosis [31]. | Prediction [6] |
| ORF7a | ORF7a is a 121-residue membrane protein plays a role in leucocyte attachment and migration. It antagonizes bone marrow stromal antigen 2 (BST-2; also known as CD317 or tetherin) which is antiviral in nature. BST-2’s attaches virions to host plasma membrane preventing further viral pathways. It also suppresses small interfering RNA which is antiviral in function [32]. |
Partially available from experiment: Residues 16-82 (PDB ID:6w37) |
| ORF7b | ORF7b is a 43-residue integral membrane protein of golgi apparatus. It is viral attenuation factor involved in human SARS-CoV-2infection [33]. | NA |
| ORF8 | ORF8 is a luminal ER membrane-associated protein with 121 residues that trigger the activation of transcription factor 6 (ATF6) and involves in human SARS-CoV-2 infection [34,35]. | 7jtletc.) |
| ORF9a (Nucleocaspid protein) | It is a 419-residue nucleocapsid protein that assembles the positive strand viral RNA genome into a helical ribonucleocapsid (RNP). This arrangement is essential during virion assembly. This protein aids in subgenomic virion RNA transcription and viral replication. This protein modulates transforming growth factor-beta signalling (TGF-β) [36]. | Partially available from experiments: Residues 41- 174 (PDB ID:6m3m [37], etc.); Residues 247-364 (PDB ID:6zco [38], etc.); |
| ORF9b | ORF9b is a97-residue protein plays a significant role in inhibition of host innate immunity by blocking a mitochondrial associated adapter. This protein favours ubiquitination and degradation of many mitochondrial dynamin-like protein [39]. | 6z4u |
| ORF9c | ORF9c is a 70-peptide present in N coding region that interacts with sigma receptors binding to lipid remodelling and evading ER stress response [40]. | NA |
| ORF10 | ORF10 a 38-residue small protein, interacts with CUL2 RING E3 ligase complex thus interfere in ubiquitination [40]. | Prediction [6] |
MATERIALS AND METHODS
Computational studies
The molecular docking studies and molecular dynamics analysis were performed using Glide Schrodinger docking software 2019_4. 3.1.
SARS-COV-2 drug targets
The following SARS-COV-2 proteins were obtained from the RSCB-Protein Data Bank using PDB codes that include: (i) the RNA dependent RNA polymerase (PDB ID: 6M71), (ii) 3CL protease (3CL pro) (PDB ID: 6M2N)
Targeting RNA-dependent RNA polymerase (6M71)
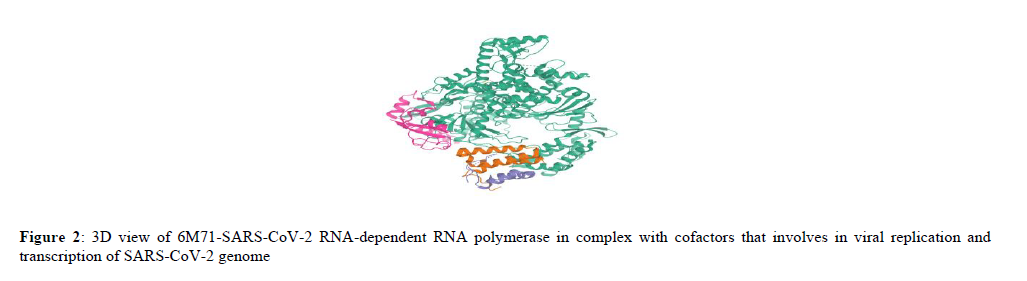
6M71 is an unligandedelectron cryo-microscopic structure of SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors and other likely assembles at 2.90 Å. The structure consists of four polypeptide chains (A, B, C, D); RNA-directed RNA polymerase (A) NSP7 (C), NSP8 (B, D) with sequence length of 942, 83, and 198 respectively. RNA-dependent RNA polymerase (RdRp) also known as NSP12 that catalyses the synthesis of viral RNA by associating with co-factors NSP7 and NSP8 thereby involves in viral replication and transcription of SARS-COV-2 genome. Currently, Remdesivir an antiviral drug in huge demand to treat moderate to severe n-Covid cases targets the viral polymerase NSP12 and hence this effort of molecular docking may shed some light on probable molecules to inhibit viral replication by interfering with NSP12 [41] (Figure 2).
Targeting 3CL protease (6M2N)
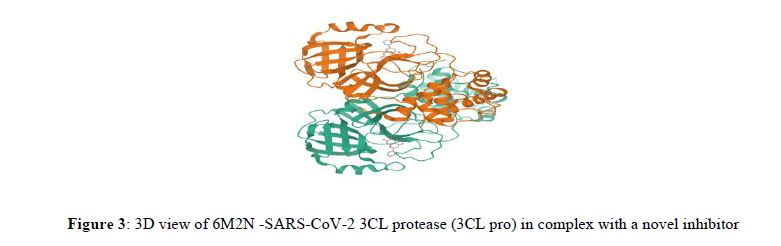
6M2N is the X-ray crystallography structure of the 3CL protease (3CL pro) in complex with 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, a novel inhibitor of SARS-CoV-2. The structure composed of four chains A, B, C, and D, each of which is identified by a single sequence-unique entity with a total of 306 amino acids. Certain proteases like 3C-like protease (3CLpro), together with a papain-like protease (PLpro), enable to transform polypeptides into mature non-structural proteins like RNA-dependent RNA polymerase (RdRp) and helicase, which are involved in essential for viral replication and maturation. These polyproteins contain 11 and 3 cleaving domains for PLpro and 3CLpro, respectively. Preferentially the substrate specificity of 3CLpro is highly conserved among different CoVs and is analogous to picornavirus 3C protease, thus rendering it an ideal target for the development of wide spectrum antiviral agents [42] (Figure 3).
Drug library
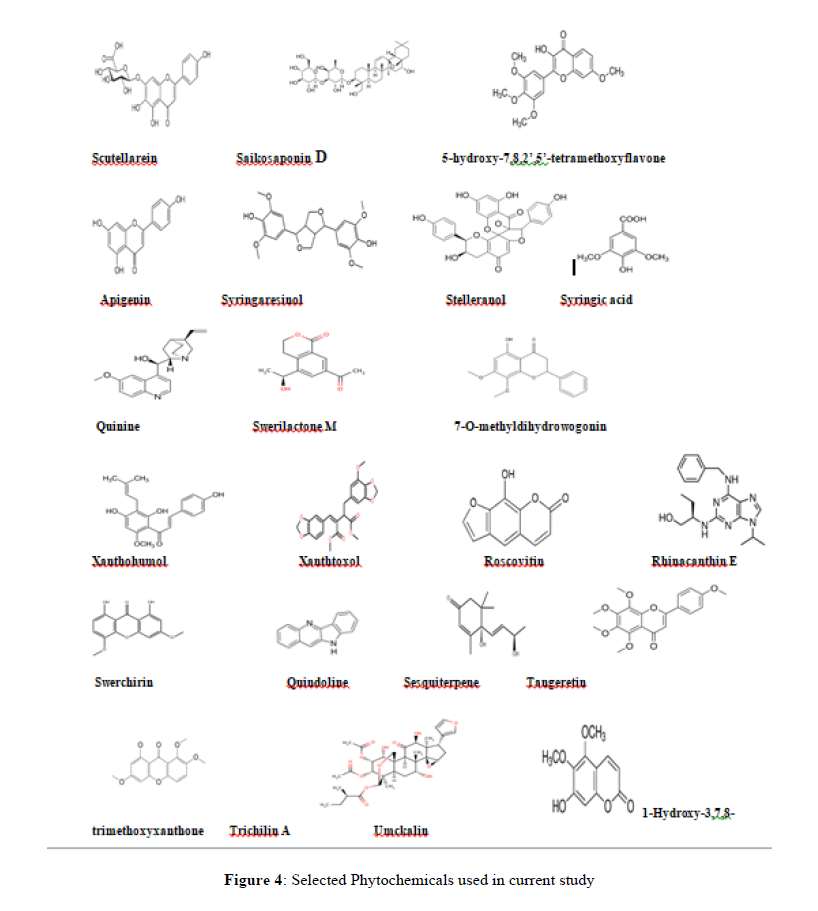
Constructed the drug library with 86 phytoconstituents for the in silico molecular virtual docking (Table 3). Selected phytoconstituents downloaded from PubChem in structure data format (SDF) format.
Validation of X-crystal proteins of drug targets
The Ramachandran two-dimensional plot was used to validate the selected protein for using them in molecular docking studies. The plot represented favoured and disfavoured torsional angles - phi (φ) and psi (ψ) of amino acids in a protein/peptide.
Protein/Target preparation
In the glide protein preparation wizard, the proteins were directly imported into the workspace by entering their PDB codes 6M71, and 6M2N. Proteins pre-processed for assigning bond orders, adding hydrogen atoms, creating disulfide bonds, etc. In the review and modify tab, we selected the required chains to generate a receptor grid, followed by soaking the co-crystallized ligand in the protein, removing water molecules, and removing small molecules. In the refine tab, these proteins were optimized and minimized to their lowest energy state and proteins with a co-crystal ligand site chosen to screen the compounds' library.
Receptor grid generation
In Maestro's task window selected the receptor grid generation to make the grid active/binding site of the protein that is suitable for docking by choosing any one atom of the co-crystal ligand molecule. It displays a grid box with X, Y, and Z coordinates. For the protein 6M2N, the co-crystals is 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, the workspace for grid formation. There is no valid co-crystal in the 3D structure of 6M71, so sitemap was used to identify the active site. As per sitemap, there were 5 active sites, and the top-ranked site was chosen for grid generation.
Ligand preparation
From the PubChem online database, imported the generated ligand library into Glide Schrodinger docking software 2019_4.3.1. Using glide ligprep, required parameters, including ionization, chirality, computation, etc., are assigned for the ligands from the workspace.
Ligand docking
Glide ligand docking was selected from tasks in Maestro, then the glide grid and ligand outmaegz zip files were loaded from the working directory. Through write XP descriptor settings, we virtually docked the compounds library.
Drug–likeness and ADME predictions
The compounds are for further evaluated for drug-likeness using SwissADME: a free web application.
RESULTS AND DISCUSSION
Molecular docking
In this study, we virtually docked phytochemical constituents against two crystal structures of SARS-CoV-2 proteins that include: (i) the RNA dependent RNA polymerase (PDB ID: 6M71), (ii) 3CL protease (3CL pro) (PDB ID: 6M2N) by using the Glide Schrodinger docking software 2019_4. 3.1.
Molecular docking of Phyto principles to RNA dependent RNA polymerase (6M71)
In Coronaviridae, NSP12 encodes for RdRp, a highly crucial enzyme involved in viral genome replication and successful transcription and hence a potential drug target. The SARS-CoV-2 NSP12 domain contains a right hand RdRP domain, and a N-terminal extension domain which is unique to nidovirus. Th amino acid sequence of polymerase domain is highly conserved in viral polymerase family with additional three subdomains viz. a finger (L366-A581 and K621-G679), a palm (residues T582–P620 and T680–Q815) and a thumb sub-domain (H816–E920). The active site is formed in the palm domain by the highly conserved polymerase motifs (A–G).
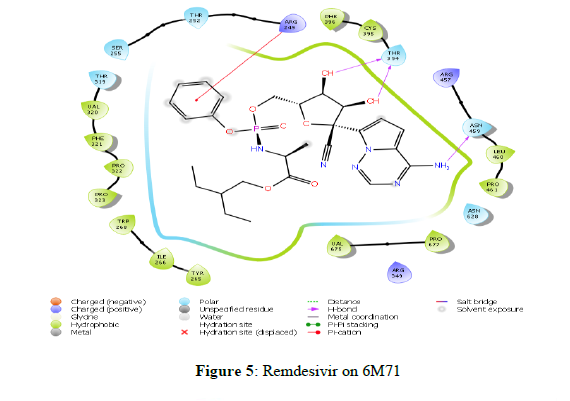
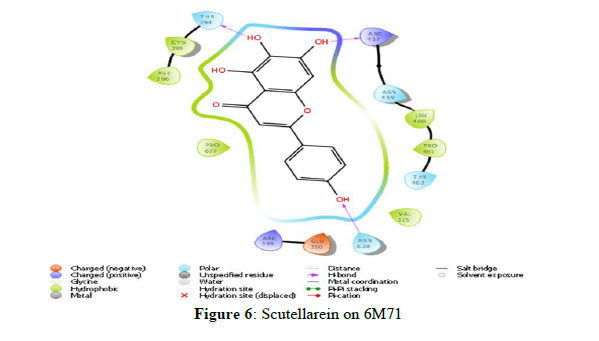
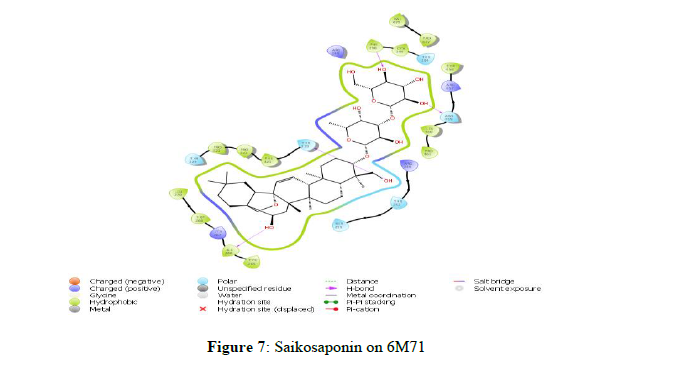
The docking scores obtained from the docking of ligands with RNA-dependent RNA polymerase (NSP12) indicated that Scutellarein and Saikosaponin D as compounds with higher binding affinity with the binding energies -39.443 and -43.588 kcal/mol respectively, and corresponding docking scores were -7.138 and -6.966. The rest of the compounds showed very weak interaction with a score of less than -4.0. However, both Scutellarein and Saikosaponin D exhibited relatively less binding interaction than the reference ligands Remdesivir (-8.767) with binding energy of -54.744 kcal/mol. The analyses of amino acid interaction of ligands on the targets indicated that binding interaction of Scutellarein (H- bond interactions with THR 394, ASN 628 and hydrophobic interaction with ARG 457) (Figure 6) and Saikosaponin D (H- bond interactions with THR 319, ILE 266, ASN 459 and hydrophobic interaction with PHE 396) (Figure 7) was quite satisfactory when compared to Remdesivir (H- bond interactions with TYR 89, GLY 165, SER 198 and hydrophobic interaction with LYS 90) (Figure 4,5) (Table 2).
Table 2: Docking results of selected Phytochemicals with RNA dependent RNA polymerase (PDB ID: 6M71)
| CHEMICAL CONSTITUENTS | PUBCHEM ID | 6M71 | |
|---|---|---|---|
| Glide score | Glide Energy | ||
| Scutellarein | 5281697 | -7.138 | -39.443 |
| Saikosaponin D | 107793 | -6.966 | -43.588 |
| 5-Hydroxy-7,8,2’,5’-tetramethoxyflavone | 10948318 | -5.967 | -35.234 |
| 5,7,2’,3-tetramethoxyflavone | 181092 | -5.676 | -42.89 |
| Apigenin | 5280443 | -5.546 | -26.596 |
| Trijugin A | 101519185 | -5.267 | -32.632 |
| Syringaresinol | 100067 | -5.041 | -37.295 |
| Stelleranol | 131676072 | -4.976 | -47.487 |
| Syringic acid | 10742 | -4.948 | -27.819 |
| Quinine | 3034034 | -4.825 | -28.909 |
| Swerilactone M | 53483971 | -4.729 | -27.836 |
| 7-O-methyldihydrowogonin | 13963770 | -4.686 | -32.67 |
| Swerilactone N | 53494394 | -4.706 | -24.053 |
| Xanthohumol | 639665 | -4.518 | -40.716 |
| Rhinacanthin E | 10366055 | -4.438 | -43.442 |
| 7-O-methylhydrowogonin | 188316 | -4.379 | -35.305 |
| Xanthtoxol | 65090 | -4.307 | -20.476 |
| Roscovitin | 160355 | -4.295 | -34.73 |
| Swerchirin | 5281660 | -4.184 | -33.049 |
| Quindoline | 98912 | -4.175 | -24.839 |
| Sesquiterpene | 6473767 | -3.712 | -24.463 |
| Schizarin B | 10582671 | -3.611 | -41.136 |
| 1-Hydroxy-3,7,8-trimethoxyxanthone | 5378284 | -3.172 | -29.101 |
| Tangeretin | 68077 | -3.11 | -36.484 |
| Trichilins A | 5462417 | -0.765 | -45.724 |
| Umckalin | 5316862 | -3.784 | -23.575 |
| Syringaldehyde | 8655 | -4.136 | -24.576 |
| Scopoletin | 5280460 | 0.167 | -26.671 |
| Skimmianine | 6760 | -3.16 | -21.724 |
| Swerilactone O | 53494395 | -3.572 | -25.33 |
| Tetrandrine | 73078 | -- | -35.216 |
| Scoparone | 8417 | -2.177 | -23.263 |
| Umbelliferone | 5281426 | -3.436 | -17.706 |
| Alpha amyrin | 73170 | -3.225 | -32.777 |
| Scopadulcic acid B | 11729855 | -2.746 | -37.686 |
| Taraxerol | 92097 | -3.193 | -36.546 |
| β-Amyrin | 73145 | -3.407 | -31.55 |
| Swertanone | 102285187 | -3.801 | -31.792 |
| Sendanin | 5352038 | -0.172 | -40.341 |
| Ursolic acid | 64945 | -3.751 | -37.243 |
| Taiwans chirin D |
70697809 | -3.385 | -37.783 |
Molecular docking of phytoprinciples to 3CL protease (3CL pro) (6M2N)
Certain proteases like 3C-like protease (3CLpro), together with a papain-like protease (PLpro), enable to transform polypeptides into mature non-structural proteins like RNA-dependent RNA polymerase (RdRp) and helicase, which are involved in essential for viral replication and maturation. These polyproteins contain 11 and 3 cleaving domains for PLpro and 3CLpro, respectively. Preferentially, the substrate specificity of 3CLpro is highly conserved among different CoVs and is analogous to picornavirus 3C protease, thus rendering it an ideal target for the development of wide spectrum antiviral agents.
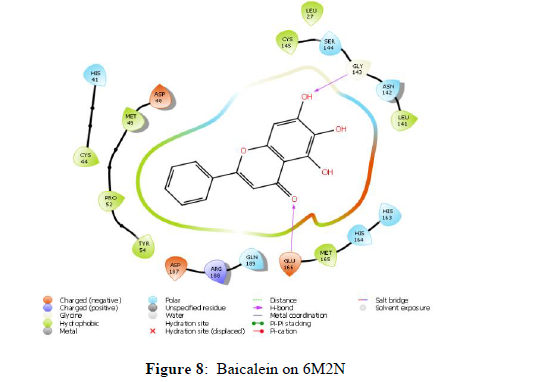
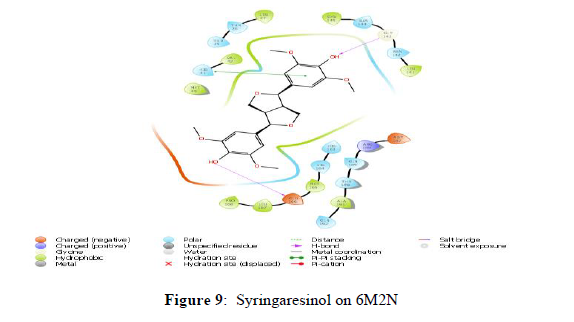
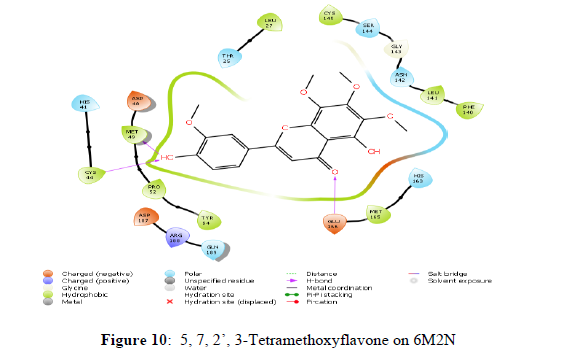
The docking scores of the docked ligands SARS-CoV-2 3CL protease (6M2N) showed that Syringaresinol and 5,7,2’,3-Tetramethoxyflavone as compounds with higher binding affinity with the binding energies -45.203 and -47.015 kcal/mol respectively, and corresponding docking scores were -7.969 and -7.453. Whilst the docking scores of co-crystals Baicalein was found to be -7.552 with a binding energy of -42.537 kcal/mol. Other constituents, showed the docking scores about -7 with the binding energy ranged from -50 to -20 (Table 3). The analyses of amino acid interaction of ligands on the targets indicated that binding interaction of Syringaresinol (H- bonding with GLY143, GLU 166) (Figure 9) and 5,7,2’,3-Tetramethoxy flavones (H- bonding with GLU 166, CYS 44, MET 49) (Figure 10) in comparison with standard Baicalein binding interactions (H-bonding with GLY 143 and GLU 166) (Figure 8) (Table 3).
Table 3: Docking results of selected Phytochemicals with 3CL protease (3CL pro) (PDB ID: 6M2N)
| Chemical constituents | Pubchem id | 6M2N | |
|---|---|---|---|
| Glide score | Glide energy | ||
| Syringaresinol | 100067 | -7.969 | -45.203 |
| 5,7,2’,3-Tetramethoxyflavone | 181092 | -7.453 | -47.015 |
| Apigenin | 5280443 | -7.421 | -41.6 |
| 7-O-Methylhydrowogonin | 188316 | -6.97 | -41.394 |
| Swerchirin | 5281660 | -6.855 | -40.713 |
| 5-Hydroxy-7,2’,6’-trimethoxyflavone | 5319878 | -6.864 | -50.801 |
| Swerilactone | 53494394 | -6.658 | -36.447 |
| Narasin | 65452 | -6.475 | -48.875 |
| 1-Hydroxy-3,7,8-trimethoxyxanthone | 5378284 | -6.414 | -39.164 |
| Roscovitin | 160355 | -6.389 | -46.466 |
ADME Predictions of Phytoprinciples Using SwissADME
Finally, compounds with good binding affinity to the selected target further analyzed the drug ability using SwissADME: a free web tool.
Results of ADME calculation
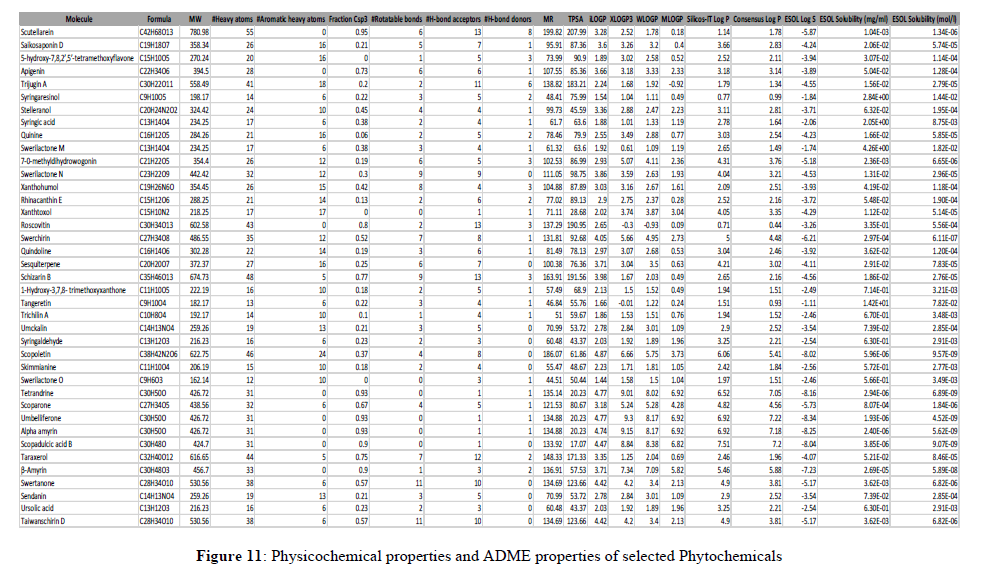
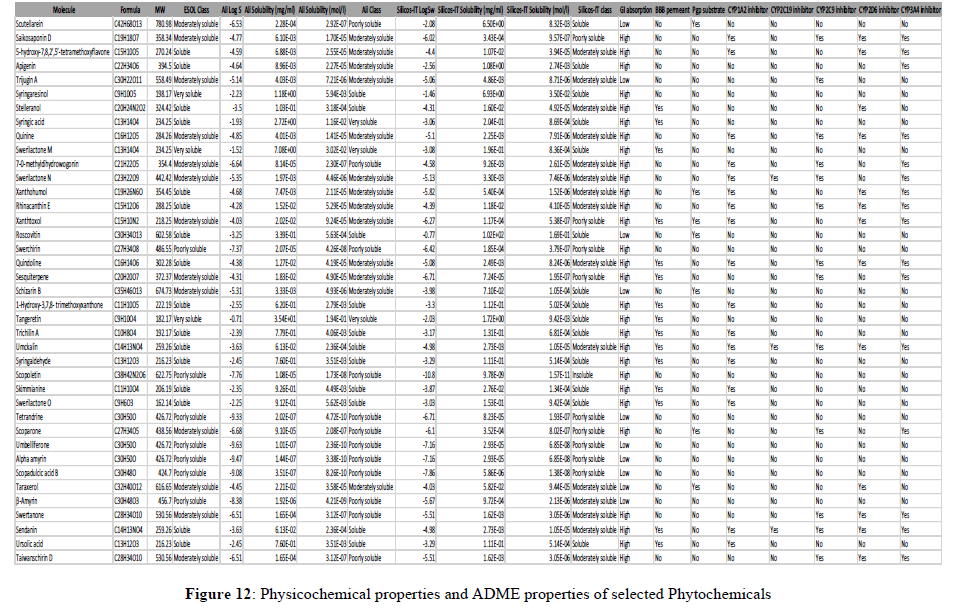
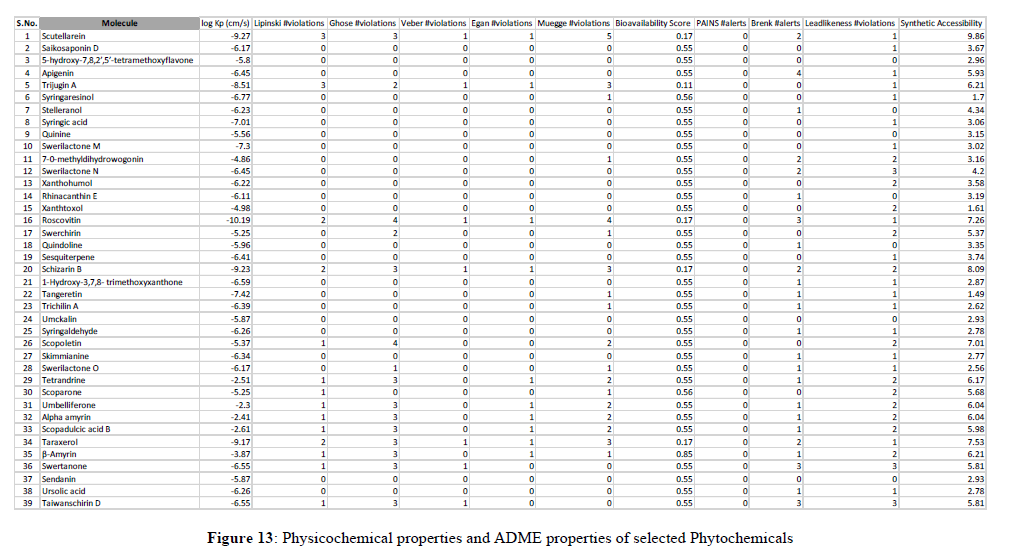
The most important and most difficult step in drug discovery and development (in which this account for the failure of about 60% of all drugs in the clinical phases) is carrying out DMPK (drug metabolism and pharmacokinetics) studies, often referred to as ADMET. In pharmacokinetic/pharmacology, ADME stands for “absorption, distribution, metabolism, and excretion”, in which they describe the disposition of a drug compound in the body. ADME Predictor is a designed program of a computer for estimating pharmacokinetic parameters/properties of drug-like compounds from their molecular structures called the ADME (Singh et al., 2013). Swiss ADME web tool is freely available software utilized to predict the physicochemical properties, absorption, distribution, metabolism, elimination and pharmacokinetic properties of molecules, which are key determinants for more clinical trials. It takes into account six physico-chemical properties, which are very vital, like lipophilicity, flexibility, saturation, polarity, solubility, and size. The result of the ADME revealed physicochemical properties of the designed compounds which includes the rules of five (MW, iLOGP, HBAs and HBDs) and several other parameters/properties like molecular polar surface area (TPSA), number of rotatable bonds (ROTBs), number of aromatic heavy atoms, and number of alerts for undesirable substructures (i.e., PAINS #alert and Brenk #alert), among others as represented in the Table 4 below. Molecular weight (MW), number of rotatable bonds (RB), number of hydrogen donors (HBD), number of hydrogen acceptors (HBA), Topo-logical Polar Surface Area (TPSA), octanol/water partition coefficient (iLOGP), number of aromatic heavy atoms (nAH), Molar refractivity(MR) and the number of alerts for undesirable substructures/sub-structures (Brenk #alert and PAINS #alert) are presented in Table 4.According to Lipinski's rule of five and the concept of QED as presented in Table 4, all the docked compounds were in accordance with the rules by causing no more than one violation. That is to say, all the MW, RB, HBD, HBA, TPSA, iLOGP, nAH and MR are within the acceptable range. Almost, 14 compounds possess a good pharmacokinetic profile with high BBB penetration presented in Figures 11-13.
SUMMARY AND CONCLUSION
In present work, the possible anti-nCOVID potential of phytochemicals were tested and analyzed through in silico methods. The docking studies pointed out the possible lead-like properties to some phytoconstituents and were validated as having drug like nature. This library was considered for tackling the modern-day issue of SARS-CoV-2 and further tested against the RNA dependent RNA polymerase (RdRp) and 3CL protease (3CL pro) key viral proteins. Analysis of protein-ligand docking revealed the following: the phytochemicals/bioactives such as Scutellarein, Saikosaponin D, Syringaresinol and 5,7,2’,3-Tetramethoxyflavone hold promise in inhibiting the SARS-CoV-2 key viral proteins. The selected phytochemicals displayed the capability to suppress SARS-CoV-2 proteins and justify their further in vitro and in vivo studies. The present study could be the starting point for the future ligands from natural sources in 2019-nCoV RdRp and 3CL pro.
References
- Ji W, Wang W, Zhao X, et al., J Med Virol. 2020, 92(4): p. 433-440.
- Wu JT, Leung K, Leung GM. The Lancet. 2020, 29: p. 689-697.
- Beura S, Chetti P. J Biomol Struct Dyn. 2020, 4: p. 1-3.
- Jauregui AR, Savalia D, Lowry VK, et al., PloS one. 2013, 8(4): p. e62416.
- Cornillez-Ty CT, Liao L, Yates JR, et al., J virol. 2009, 83(19): p. 10314-10318.
- Z. Lab. Genome-wide structure and function modeling of SARS-CoV 2.
- Báez-Santos YM, John SE, Mesecar AD. Antiviral Res. 2015, 115: p. 21-38.
- Michalska K, Kim Y, Jedrzejczak R, et al., IUCrJ. 2020, 7(5): p. 814–824.
- Sakai Y, Kawachi K, Terada Y, et al., Virology. 2017, 1: p. 165-174.
- Jumper J, Tunyasuvunakool K, Kohli P, et al., Computational predictions of protein structures associated with COVID-19. 2020, 5: p. 2.
- Wu C, Liu Y, Yang Y, et al., Acta Pharmaceutica Sinica B. 2020, 10(5): p.766-88.
- Douangamath A, Fearon D, Gehrtz P, et al., Nat Commun. 2020, 11(1): p. 1-1.
- Angelini MM, Akhlaghpour M, Neuman BW, et al., MBio. 2013, 4(4): p. e00524-13
- Cottam EM, Whelband MC, Wileman T. Autophagy. 2014, 10(8): p. 1426-1441.
- Kirchdoerfer RN, Ward AB. Nat Commun. 2019, 10(1): p. 1-9.
- Snijder EJ, Decroly E, Ziebuhr J. Adv Virus Res. 2016, 96: p. 59-126.
- Rogstam A, Nyblom M, Christensen S, et al., Int J Mol Sci. 2020, 21(19): p. 7375.
- Gao Y, Yan L, Huang Y, et al., Science. 2020, 368(6492): p. 779-782.
- Jang KJ, Jeong S, Kang DY, et al., Sci Rep. 2020, 10(1): p. 1-3.
- Ferron F, Subissi L, De Morais AT, et al., Proc Natl Acad Sci. 2018, 115(2): p. E162-171.
- Deng X, Hackbart M, Mettelman RC, et al., Proc Natl Acad Sci. 2017, 114(21): p. E4251-4260.
- Rosas-Lemus M, Minasov G, Shuvalova L, et al., bioRxiv. 2020, Apr 26.
- Hoffmann M, Kleine-Weber H, Schroeder S, et al., cell. 2020, 181(2): p. 271-280.
- Chi X, Yan R, Zhang J, et al., Science. 2020, 369(6504): p. 650-655.
- Siu KL, Yuen KS, Castano‐Rodriguez C, et al., The FASEB Journal. 2019, 33(8): p. 8865-8877.
- Kern DM, Sorum B, Mali SS, et al., BioRxiv. 2020, 28(7): p. 573-582.
- Kamau A, Kulmanov M, Arold ST, et al., bioRxiv. 2020.
- Schoeman D, Fielding BC. Virol J. 2019, 16(1): p. 1-22.
- Mandala VS, McKay MJ, Shcherbakov AA, et al., Nat Struct Mol Biol. 2020, 27(12): p. 1202-1208.
- Voß D, Pfefferle S, Drosten C, et al., Virol J. 2009, 6(1): p. 1-3.
- Huang SH, Lee TY, Lin YJ, et al., J Microbiol Immunol Infect. 2017, 50(3): p. 277-285.
- Taylor JK, Coleman CM, Postel S, J Virol. 2015, 89(23): p. 11820-33.
- Pfefferle S, Krähling V, Ditt V, et al., Virol J. 2009, 6(1): p. 1-7.
- Muth D, Corman VM, Roth H, et al., Scientific reports. 2018, 8(1): p. 1-1.
- Sung SC, Chao CY, Jeng KS, et al., Virology. 2009, 387(2): p. 402-413.
- Mu J, Xu J, Zhang L, et al., Sci China Life Sci. 2020, 63(9): p. 1-4.
- Kang S, Yang M, Hong Z, et al., Acta Pharm Sin B. 2020, 10(7): p. 1228-1238.
- Zinzula L, Basquin J, Bohn S, et al., Biochem Biophys Res Commun. 2021, 538: p. 54-62.
- Shi CS, Qi HY, Boularan C, et al., J Immunol. 2014, 193(6): p. 3080-3089.
- Gordon DE, Jang GM, Bouhaddou M, et al., Nature. 2020, 583(7816): p. 459-68.
- Gao Y, Yan L, Huang Y, et al., Science. 2020, 368(6492): p. 779-82.
- Su HX, Yao S, Zhao WF, et al., Acta Pharmacol Sin. 2020, 41(9): p. 1167-77.
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref
Indexed at, Google Scholar , Crossref