Research - Der Pharma Chemica ( 2021) Volume 13, Issue 9
In-Silico Prediction of T- cell epitopes in VacA: First Step towards the Design of a Helicobacter Pylori Vaccine
Elijah Kolawole Oladipo1,2*, Abisola Temiloluwa Aina1, Oluwaseyi Febenna Eko1, Olubukola Monisola Oyawoye1, Boluwatife Ayobami Irewolede2 and Temitope Isaac Adelusi32Genomics Unit, Helix Biogen Institute, Ogbomoso, Oyo State, Nigeria
3Computational Biology/Drug Discovery Laboratory, Department of Biochemistry, Ladoke Akintola University of Technology, P.M.B. 4000, Ogbomoso, Nigeria
Elijah Kolawole Oladipo, Department of Microbiology, Laboratory of Molecular Biology, Immunology and Bioinformatics, Adeleke University, P.M.B. 250, Ede, Osun State, Nigeria, Email: koladipo2k3@yahoo.co.uk
Received: 23-Aug-2021 Accepted Date: Sep 24, 2021 ; Published: 30-Sep-2021
Abstract
Helicobacter pylori are indicated in gastritis, gastric cancer, Peptic ulcer and B cell mucosa lymphoid tissue lymphoma. The bacteria have a 50% pandemic, a higher prevalence of 75% in developing countries and 40% in developed countries. A combination of antibiotics is used to treat Helicobacter pylori infections. Some of these are Amoxicillin, tetracycline, and fluoroquinolones, but are ineffective in eliminating the infections and are also compromised by antibiotic-resistant strains, therefore a vaccine to help prevent, control and eliminate Helicobacter pylori infections is needed. We proposed to predict T-cell epitopes of VacA. Seven VacA protein sequences were retrieved from the NCBI protein database and subjected to antigenicity test using the ANTIGENpro database. All the seven (7) sequences with antigenicity score ≥0.8 were submitted to the NetCTL server and IEDB (Immune Epitope Database) MHC-II server to predict the cytotoxic T-lymphocyte (CTL) and to predict the Helper T-lymphocyte (HTL) epitopes, respectively. The cytotoxic T-lymphocyte epitopes and Helper T-lymphocyte epitopes were predicted. The seven amino acid sequences obtained from NCBI subjected to antigenicity test passed by having a score of ≥0.8. Ninety-eight (98) CTL (9-mer) ligands in total were predicted for the seven selected proteins submitted to NetCTL sever. Fifty-six (56) epitopes were selected according to their high scores. Ninety-nine (99) (15-mer) total HTL epitopes were predicted. Their IC50 value was not greater than 500nm with percentile rank ranging from 0.030-4.00, while ninety-four (94) epitopes were selected. A total of fifty-six (56) CTL and Ninety-four (94) HTL epitopes were merged using AAY, EAAK, and GPGPG linkers with sequence APPHALS as an adjuvant to construct the multi-epitope vaccine. The predicted peptides are presumed to be valuable in the multi-epitope vaccines, non-allergen without compromising the population coverage. VacA has potential B-cell and T-cell epitopes distributed throughout the sequence. Thus several fragments were identified as valuable candidates for subunit vaccines against Helicobacter pylori.
Keywords
Helicobacter pylori, Peptic Ulcer, Vaccine, Gastric cancer, VacA
Introduction
Helicobacter pylori are gram-negative, microaerophilic bacteria. They are spiral shaped flagellates and populate the human gastric mucus membrane [1]. H. pylori colonization typically does not trigger clinical presentation but can increase the risk of developing gastrointestinal ulcer diseases, lymphoma, and gastric cancer. H. pylori infections in many patients are asymptomatic, and only a few people develop clinical manifestations [2]. Helicobacter pylori are responsible for Gastritis, Gastric cancer, Peptic ulcer, B cell mucosa lymphoid tissue lymphoma [3]. The bacteria are 50% pandemic and have a higher prevalence of 75% in developing countries but, in developed countries, prevalence is 40% [1]. The virulence factor of Helicobacter pylori are vacA, cagA, dupA, oipA, and s1m1. Infection by H. pylori with positive vacA, s1m1and the cagA genes increases the possibility of developing gastric cancer. The correlation between vacA, s1m1, and the cagA and gastric cancer indicates that screening of these genes may be beneficial for detecting populations at greater risk for gastric cancer [4,5]. Co-expressing H. pylori virulence factors can predict severe H. pylori infection, like a peptic ulcer or gastric cancer. Thus far, several new virulence factors of H. pylori have been depicted; nonetheless, an established correlation between these new virulence factors and gastric cancer has not been presented; however, it has been projected that specific analysis of H. pylori virulence factors can be pertinent for predicting post-H. Pylori infection disorders [6,7]. Over the years, Helicobacter pylori infections have been treated with a combination of antibiotics, some of which are Amoxicillin, tetracycline, and fluoroquinolones but are not effective enough to eliminate the infection and are also compromised by antibiotic-resistant strains [8] hence a vaccine to help prevent, control and eliminate Helicobacter pylori infections is needed. Epidemiological surveys indicate that up to 90% of gastric cancer could be attached to Helicobacter pylori infection and that elimination of this pathogen could relegate the prevalence of gastric cancer. Screening for and suppression of H. pylori is consequently an insinuating approach to eradicate gastric cancer. This study suggests the design of a multi-epitope vaccine against Helicobacter pylori infections.
Materials and Methods
Retrieval of protein sequences and antigenicity testing
Seven VacA protein sequences were retrieved in FASTA format with accession number AAU85846.1, ADPO2392.1, ADPO2391.1, ADPO2390.1, ADK63389.1, ADK63388.1, and ADK63387.1 from the NCBI protein database after which their antigenicity was predicted using the ANTIGENpro database (http://scratch.proteomics.ics.uci.edu/). All the protein sequences selected obtained a probability for antigenicity score of ≥ 0.8 [9].
Prediction of CTL Epitopes
All the seven (7) sequences with antigenicity score ≥ 0.8 were subjected to NetCTL (http://www.cbs.dtu.dk/services/NetCTL/) in FASTA format. The cytotoxic T-lymphocyte epitope was predicted at the default threshold score of 0.75, those with a combined score >0.75 were further selected as CTL epitopes and IEDB MHC I immunogenicity model was used to predict their HTL epitopes [9].
Prediction of HTL Epitopes
VacA protein sequences retrieved from NCBI server are subjected to Immune Epitope Database (IEDB) MHC-II epitope prediction module (http://www.iedb.org/). The selected alleles for prediction are H2-IAb, H2-IAd and H2-IEd mouse alleles, while other parameters were left default.
Multi-epitope subunit vaccine sequence construction
A total of 56 CTL and 94 HTL epitopes were merged together using AAY and GPGPG linkers. AAY linker was used to bind intra-CTL epitopes and a GPGP linker to bind intra-HTL epitopes while using a TLR-4 agonist with sequence APPHALS as an adjuvant. APPHALS was joined to the CTL and HTL epitopes by EAAY linkers.
Vaccine allergenicity and antigenicity prediction
Antigenicity of the constructed multi-epitope subunit vaccine was predicted using ANTIGENpro sever (http://scratch.proteomics.ics.uci.edu). ANTIGENpro utilizes protein antigenicity microarray data to predict protein antigenicity. This server is an alignment-free and pathogen-independent predictor that is sequence-based [9] while AllerTOP v. 2.0 (https://www.ddg-pharmfac.net/AllerTOP/) online servers was used to predict the allergenicity.
Determination of physiochemical properties
Physiochemical properties such as amino acid composition, theoretical pI, instability index, in vitro and in vivo half-life, aliphatic index and molecular weight were predicted with ProtParam sever (http://web.expasy.org/protparam/).
Results
Retrieval of protein sequences and antigenicity testing
Table 1 shows the seven amino acid sequences with accession numbers AAU85846.1, ADPO2392.1, ADPO2391.1, ADPO2390.1, ADK63389.1, ADK63388.1, and ADK63387.1 obtained from the NCBI database in FASTA format, which was selected according to their antigenicity score obtained from the ANTIGENpro server. All the proteins have a score of ≥ 0.8 [9].
| S/N | Protein Accession No. | Antigenicity Score | Selected/Non Selected |
|---|---|---|---|
| 1 | AAU85846.1 | 0.9590 | Selected |
| 2 | ADPO2392.1 | 0.9581 | Selected |
| 3 | ADPO2391.1 | 0.9581 | Selected |
| 4 | ADPO2390.1 | 0.9557 | Selected |
| 5 | ADK63389.1 | 0.8728 | Selected |
| 6 | ADK63388.1 | 0.8983 | Selected |
| 7 | ADK63387.1 | 0.9126 | Selected |
Cytotoxic T-lymphocyte (CTL) Epitope Prediction and Immunogenicity Assessment
98 CTL (9-mer) ligands in total were predicted for the seven selected proteins, and 56 epitopes were selected according to their high scores, as shown in Table 2.
| S/N | Protein Accession No. | Epitopes | Comb. score | Length | IEDB immunogenicity score | Selected/Non-selected |
|---|---|---|---|---|---|---|
| 1 | AAU85846.1 | DMKDAVGTY SKNAEISLY YSTINTSKV NTSKVTGEV KTEIQPTQV LTTNAAHLH TTNAAHLHI RVNNQVGGY GTDTKNGTA GTATFNNDI KVDAHTANF GIDTGNGGF VTDKVNINK YSQFSNLTI DIDSATGFY LLKAKIIGY |
0.9988 0.9016 0.8096 1.1167 0.7704 0.8141 0.9979 1.2065 1.3160 0.9499 0.8680 0.8486 1.3597 0.9710 2.9331 0.7576 |
9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 |
0.08056 0.11395 -0.08886 -0.10686 0.00826 0.10184 0.11536 -0.03415 -0.11855 0.16717 0.14385 0.12157 -0.06715 -0.07367 0.01237 0.01677 |
Selected Selected Non-selected Selected Selected Selected Non-selected Non-selected Selected Selected Selected Non-selected Non-selected Selected Selected Selected |
| 2 | ADPO2392.1 | DMKDAVGTY ITSDKNAEI TSDKNAEIS YSTINTSKV NTSKVTGEV QSSNSNTEV KTEIQPTQV TTNAAHLNI RVNNQVGGY GTATFNNDI KVDAHTANF GIDTGNGGF YSQFSNLTI DIDSATGFY LLKAKIIGY SMVNNPDNY RSKDIDTLY LIDSHDAGY ATDALNNVA |
0.9989 0.7744 0.7895 0.8096 1.1167 1.2607 0.7620 0.7918 1.2088 0.9499 0.8680 0.8486 0.9710 2.9331 0.7576 0.8944 1.9109 2.4031 1.2167 |
9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 |
0.08056 -0.15595 -0.01701 -0.08886 -0.10686 -0.13614 0.00826 0.09268 0.12908 0.16717 0.14385 0.12157 -0.07367 0.01237 0.01677 0.00509 0.12908 -0.05407 0.04834 |
Selected Non-selected Non-selected Non-selected Non-selected Selected Selected Selected Selected Selected Non- selected Selected Selected Selected Non-selected Non-selected Non-selected Non-selected Non-selected |
| 3 | ADPO2391.1 | DMKDAVGTY YSTINTSKV NTSKVAGEV KTEIQPTQV LTTNAAHLH TTNAAHLHI MVNNQVSGY GTDTNNGTI DTNNGTIAF DIDTGNGGF KLVINDFYY LLKAKIIGY SMVNNPDNY RSSDIDTLY SSDIDTLYA LIDSHNAGY NIDSFAQRL GTSAGVDAY GSDQSSLNF LLRDLNQSY NQSYNYLAY YSAATRASY AATRASYGY ASANVEARY SANVEARYY YYGDTSYFY FASNLGMRY |
0.9989 0.8283 1.0111 0.7686 0.8141 0.9979 1.7462 1.9771 0.8203 0.8367 0.7683 0.7572 0.8944 3.0555 1.4235 2.8021 0.7645 2.6876 2.3621 0.9202 1.9312 2.5136 0.8299 2.1570 1.0905 1.0136 2.4986 |
9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 |
0.08056 -0.08886 -0.10657 0.00826 0.10184 0.11536 -0.20237 0.08515 0.19611 0.12157 0.25854 0.01677 0.00509 0.14538 0.18774 -0.08104 -0.07596 0.09911 0.08515 -0.17219 -0.0537 0.07495 -0.03627 0.2039 0.21529 -0.01933 -0.15707 |
Selected Non-selected Non-selected Selected Selected Non-selected Selected Selected Selected Selected Selected Selected Selected Selected Non-selected Selected Selected Non-selected Non-selected Selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected |
| 4 | ADPO2390.1 | DMKDAVGTY YSTINTSKV NTSKVTGEV KTEIQPTQV TTNAANLNI MVNNQVGGY GVDTKNGTI DIDTGNGGF KLVINDFYY YSQFSNLTI NVDSATGFY LLKAKIIGY SMVNNPDNY TSNARFASY RSSDIDTLY SSDIDTLY LIDSHDAGY NSGGNTSLY GTSAGVDAY GSDQSSLNF LLRDLNQSY NQSYNYLSY ASTRASYGY ASANVEARY SANVEARYY YYGDTSYFY FASNLGMRY |
0.9989 0.8096 1.1167 0.7686 0.8941 1.7930 0.8543 0.8367 0.7683 0.9567 3.2718 0.7572 0.8944 2.2171 3.0554 1.4101 2.4054 2.4453 2.6964 2.3621 0.9213 1.7050 1.4402 2.1570 1.0905 1.0136 2.4986 |
9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 |
0.08056 -0.08886 -0.10686 0.00826 0.05614 -0.03415 -0.11855 0.12157 0.25854 -0.07367 0.01237 0.01677 0.00509 0.13423 0.14538 0.18774 -0.05407 -0.07076 0.09911 -0.43933 -0.17219 -0.17322 -0.03627 0.2039 0.21529 -0.01933 -0.15707 |
Selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected Non-selected |
| 5 | ADK63389.1 | DMKDAVGTY ITSDKNAEI TSDKNAEIS |
0.9988 0.7744 0.7895 |
9 9 9 |
0.08056 -0.15595 -0.01701 |
Selected Non-selected Non-selected |
| 6 | ADK63388.1 | DMKDAVGTY | 0.9988 | 9 | 0.08056 | Selected |
| 7 | ADK63387.1 | DMKDAVGTY | 0.9989 | 9 | 0.08056 | Selected |
Helper T-lymphocyte (HTL) Epitope Prediction
IEDB MHC-II prediction tool was further used to predict the MHC-II binding molecules of the six epitopes. A total of 27 HTL epitopes were predicted. Their IC50 value ranging from 1284 – 5366 with percentile rank ranging from 15-16 (Table 3).
| Seq. no | Alelle | Start | End | Epitope | Percentile Rank | IC50 | Method Used |
|---|---|---|---|---|---|---|---|
| 1 | H2-IAb | 240 | 254 | DGATLNLASSSVKLM | 15.00 | 2054.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 40 | 54 | IIPAIVGGIATGAAV | 15.00 | 2041.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 267 | 281 | AYLAPSYSTINTSKV | 15.00 | 2586.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 155 | 169 | DLDVNMQKATLRLGQ | 15.00 | 2642.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 241 | 255 | GATLNLASSSVKLMG | 15.00 | 3399.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 531 | 545 | NINKLITASTNVAIK | 15.00 | 3596.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 706 | 720 | ATGFYKPLIKINSAQ | 15.05 | 3161.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 295 | 309 | HNAAQAGIIASNKTH | 15.05 | 5323.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 216 | 230 | SSTVLTLQASEGITS | 15.05 | 5366.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 98 | 112 | YDLYKSLLSSKIDGG | 15.15 | 2341.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 289 | 309 | HLTVGDHNAAQAGII | 15,20 | 1284.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 270 | 284 | APSYSTINTSKVTGE | 15.25 | 2049.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 534 | 548 | KLITASTNVAIKNFN | 15.50 | 2571.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 17 | 31 | LALVGALVSITPQQS | 15,50 | 2268.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 31 | 45 | SHAAFFTTVIIPAIV | 15.50 | 1929.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 242 | 256 | ATLNLASSSVKLMGN | 15.50 | 3563.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 532 | 546 | INKLITASTNVAIKN | 15.50 | 3414.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 543 | 547 | NKLITASTNVAIKNF | 15.50 | 3624.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 377 | 391 | TQVINGPFAGGKDTV | 15.65 | 3063.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 405 | 415 | ADGTIRVGGYKASLT | 15.75 | 4843.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 28 | 42 | PQQSHAAFFTTVIIP | 16.00 | 1735.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 378 | 392 | QVINGPFAGGKDTVV | 16.00 | 2421.00 | Consensus (smm/nn) |
| 1 | H2-IAb | 27 | 41 | TPQQSHAAFFTTVII | 16.00 | 1764.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 570 | 584 | DIGSQSRINTVRLET | 16.00 | 2895.00 | Consensus (smm/nn) |
| 1 | H2-IAd | 15 | 29 | VSLALVGALVSITPQ | 16.00 | 3961.00 | Consensus (smm/nn) |
| 1 | H2-IEd | 723 | 727 | IKNTEHVLLKAKIIG | 16.00 | 4815.00 | Consensus (smm/nn) |
Construction of multi-epitope vaccine
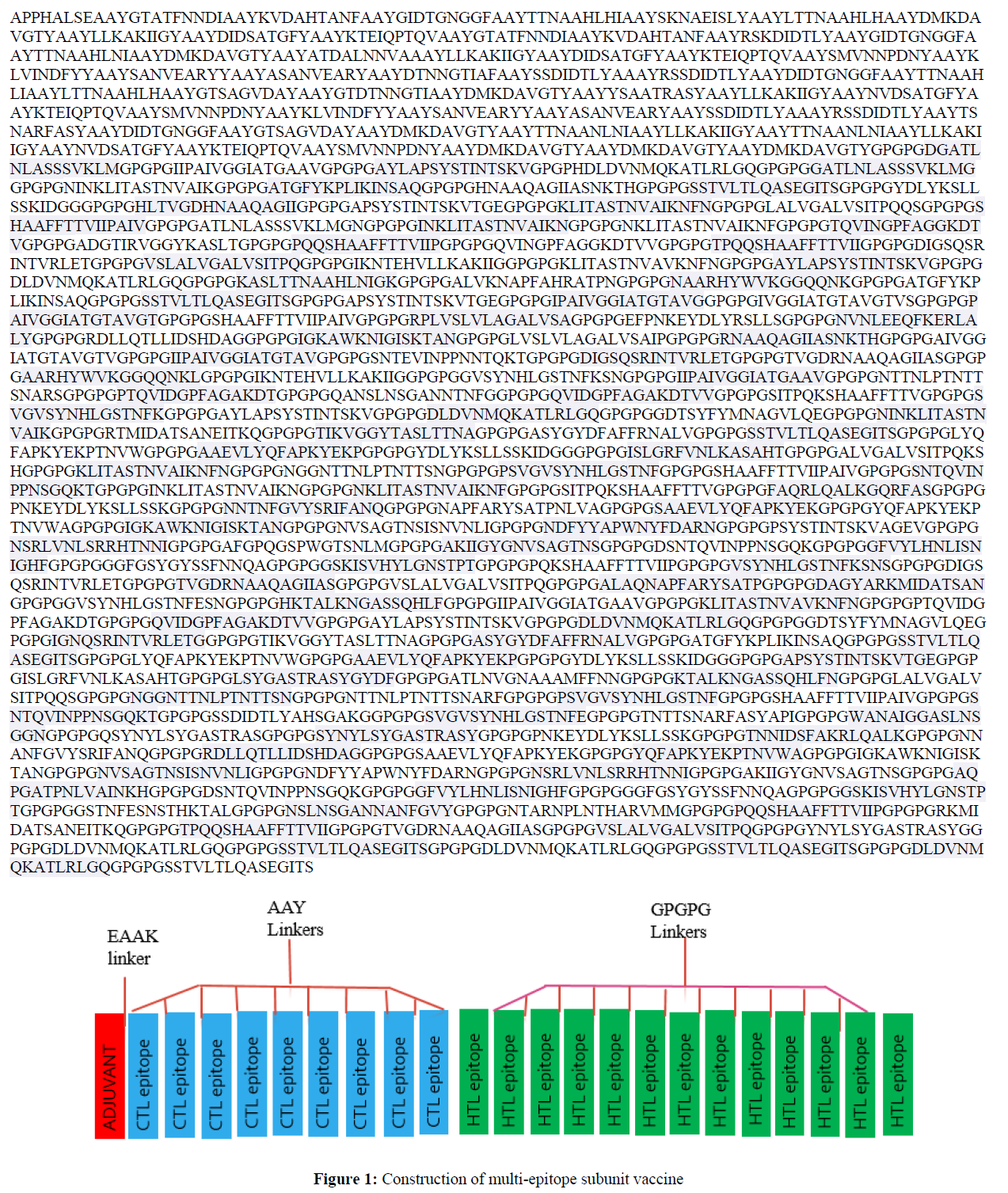
This vaccine was constructed with the 56 CTL and 27 HTL epitopes consisting of amino acid residues and an Adjuvant. The EAAK linker linked CTL epitopes, intra-CTL and Intra-HTL epitopes were linked by AAY and GPGPG linker, respectively. The final vaccine construct has a suitable adjuvant, linker, and immunogenic CTL, and HTL epitopes moving from N-terminal to C-terminal.
Construction of multi-epitope subunit vaccine (Figure 1)
Vaccine allergenicity and antigenicity prediction
AllerTOP online server was used to determine the allergenicity of the predicted vaccine construct; it was found that the vaccine protein is non-allergen in nature and safe for human use. The antigenicity of the designed vaccine construct was determined by the ANTIGENpro server.
Determination of physicochemical properties
The molecular weight (MW) of the final protein was predicted to be 407329.10 Da with the formula: C18146H27908N5008O5636S29. The protein is predicted to be alkaline based on the theoretical isoelectric point value (pI) of 9.32. The half-life was assessed to be 4.4 hours in mammalian reticulocytes in vitro, > 20 hours in yeast in vivo and > 10 hours in Escherichia coli in vivo. An instability index (II) of 14. Seventy-five classifying the protein as stable. Thermostability was indicated by the estimated aliphatic index predicted to be 67.26, while the predicted Grand average of hydropathicity (GRAVY) was -0.234.
Discussion
Vaccination is one of the utmost applicable processes to proficiently, precipitously, and reasonably advance public health and the preeminent way to constrain infectious disease in society [9]. Up until now, exertions to expound a suitable all-inclusive vaccine against Helicobacter pylori bacterial infections have been ineffective. Recent research is centered on subunit vaccines as compared to whole organism vaccines because subunit vaccines contain distinctive immunogenic constituents of the pathogens liable for the infection rather than the whole pathogenic agent [9]. Thus, in this study, the major focus was detecting conserved antigenic epitopes in selected VacA protein sequences.
Different criteria were consecutively exploited to recognize the most suitable epitopes for our vaccine design, and selected T-cell epitopes, which are antigenic and capable of inducing antibody production, were detected. CTL and HTL epitope prediction was carried out base on their need in a vaccine construct to help elicit an effective adaptive immunity for successful protection against pathogen infection [10,11]. However, CTL plays a cardinal role in the battle against infections. Therefore, to a great extent, various studies have elucidated the technique behind its response to microorganisms [12]. Degraded proteins of microorganisms found on the surface of infected cells are presented to the CTLs in combination with the MHC Class I molecules. Upon recognizing the degraded part of such proteins by CTLs, cytotoxic granules are released, thus leading to the death of the infected cells [12]. Filtering through various parameters, 56 CTL epitopes have been extracted in this study for the construction of the candidate vaccine.
Following the cleavage of specific antigens to body cells, antigen-presenting cells readily present the epitopes to HTLs in the form of epitope-MHC class II complex. This interaction leads to the activation of HTLs, which thereby result in the secretion of a wide range of cytokines and chemokines such as IFNγ, IL-4, IL-10 etc. that play diverse roles in the immune response against invaders upon recognition [12]. Based on the analysis from this study, 94 HTL epitope was selected for the vaccine construction. The detected epitopes entail experimental validation for their possible use in peptide vaccines. Since immune informatics is a field that can predict a priori whether an antigen is immunogenic through epitope prediction, our focus was to stage in silico prediction of T-cell epitopes within the amino acid sequence of VacA for the design of a subunit vaccine against H. pylori. A passable humoral response is a contingent upon an appropriate CD4+ T-cell response.
Furthermore, vaccines that induce cellular immune responses are crucial for infections initiated by intracellular pathogens. One of the main drawbacks in developing such vaccines includes obscurity in recognizing relevant T-cell antigens that stimulate protective cellular immunity [10]. T-cells detect the peptide epitopes posed by the MHC molecules, which are the surface proteins of antigen-presenting cells recognized by the T-cell receptors (TCR). These antigen-presenting molecules (MHC) are either class I or II. MHC class I molecules pose on nearly all nucleated cells that specifically signify the endogenous proteins or antigen processed through the cytosolic pathway and denotes cytotoxic T lymphocytes (CTLs). MHC class II molecules pose exogenous antigens, ordinarily, surface proteins of the pathogens, which are processed through endocytic pathways and accessible to helper T lymphocytes or CD4+ T cells [9]. In this study, novel immune-informatics tools were exploited to design a prospective, benign and immunogenic subunit vaccine that may have the proficiency to constrain Helicobacter pylori infections.
Conclusion
VacA has probable B-cell, and T-cell epitopes dispersed all over the whole sequence. Various fragments could be designated as candidates for the design of a vaccine against Helicobacter pylori. Consequently, in this study, seven sequences comprising both B-cell and T-cell epitopes were distinguished that may be used to spawn both humoral and cellular responses. These antigenic peptides can be significant candidates for indicative, curative, and preventive vaccines against H. pylori. Selected fragments of VacA that contains B- and T-cell epitopes in conjunction with effective adjuvants and/or delivery systems may facilitate the enhancement of an effective H. pylori vaccine that prompts a Th1-mediated and humoral protective immune response that does not instigate antagonistic immune-pathologies. The results of our study provide analytical data for the detection and assessment of epitopes in VacA and may be exploited for the development of epitope vaccines that have improved safety and efficacy.
References
- Safavi M, Sabourian R and Alireza F. World J Clin Cases. 2016, 4(1): p. 5-19.
- Goudarzi, Mehdi, Heidary et al., Gastroenterol Hepatol Bed Bench. 2016, 9(1). S47–S52.
- Testerman TL and James Morris. World J Gastroenterol. 2014, 20(36): p. 12781-12808.
- Sallas LM, Melchiades LJ, Zabaglia ML et al., Adv Appl Microbiol. 2017, 7(1): p. 1-9.
- Pormohammad Ali, Ghotaslou Reza, Leylabadlo et al., Microb. Pathog. 2018, 118: p. 214-219.
- Matos Joana, De Sousa, Henrique et al., Eur J Gastroenterol Hepatol. 2013, 25(12): p. 1431-1441.
- Abadi ATB, Mobarez AM, Bonten MJ et al., BMC Gastroenterol. 2014, 14(33).
- Garza-Gonzalez E, Perez-Perez, Guillermo et al., World J Gastroenterol. 2014, 20(6): p. 1438-1449.
- Khatoon N, Pandey RK and Prajapati VK. Scientific Reports. 2017, 7(8285).
- Karunakaran KP, Yu H, Foster LJ et al., Hum Vaccine. 2010, 6(8): p. 676-680.
- Bettini E and Locci M. Vaccines. 2021, 9: p. 147.
- Ishtiaque Ahammad and Samia Sultana Li. J. Biol. Macromol. 2020.