Research - Der Pharma Chemica ( 2020) Volume 12, Issue 5
Predicted Treatment for COVID-19 Infection: Design, Docking Study and ADMET Prediction of Novel Small Molecules as Serine Protease Inhibitors
Phoebe F. Lamie* and John N. PhiloppesPhoebe F. Lamie, Department of Pharmaceutical Organic Chemistry, Faculty of Pharmacy, Beni-Suef University, Beni-Suef 62514, Egypt, Email: feby.farag@yahoo.com
Received: 13-Aug-2020 Accepted Date: Aug 24, 2020 ; Published: 28-Aug-2020
Abstract
Nowadays, pandemic coronavirus disease COVID-19, first appeared in Wuhan, China, in December 2019, results in serious global threats to public health, economic and social habits. The aim of this work is to design new compounds with general pharmacophores that mimic seine protease inhibitors (such as camostat mesylate), TMPRSS2 inhibitors, antiviral agents and mucolytic cough suppressant agents. The designed eight compounds were subject to molecular docking study, drug-likeness and in-silico ADMET prediction. Most of the compounds, especially TDa, TDd, TDe, TDf and TDh exhibited good results in docking study inside TMPRSS2 active site and bioavailiability and toxicity evaluation. They could be promising candidates as drug leads for development anti-SARS-Cov-2, the fatal pandemic disease, and therapeutics in the near future.
Keywords
COVID-19, Docking study, ADMET, TMPRSS2, Serine protease inhibitors
Introduction
From highly pathogenic viruses that have arisen in the last 40 years, Ebola viruses, Sever Acute Respiratory Syndrome (SARS-coV) and Middle East Respiratory Syndrom (MERS-coV) which appeared in 2012 [1-3]. Until now, there is no vaccines or therapeutic drugs approved from FDA, and their treatments depends on drug overlapping using broad spectrum antiviral drugs [4].
Nowadays, pandemic coronavirus disease COVID-19, first appeared in Wuhan, China, in December 2019, results in serious global threats to public health, economic and social habits [5-7].
COVID-19 (SARS-coV-2), causes acute infection to respiratory system, especially lungs leading to symptoms such as fever, tiredness, dry cough, nasal congestion, sore throat, difficulty breathing and diarrhea [8-10].
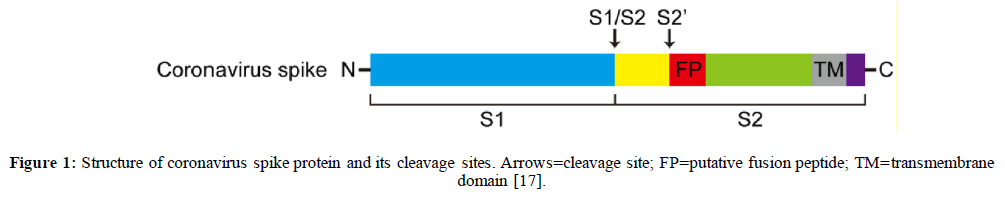
COVID-19 is an enveloped virus with a single positive strand RNA genome (+SSRNA). This envelop is covered by a glycosylated fusion spike (S) protein [11-13]. The s-protein consists of two functional domains, S1 and S2 subunits. The receptor binding domain (RBD), S1, is responsible for virus binding to the cell surface receptor, while, S2 subunit contains functional elements involved in membrane fusion [14-16], Figure 1.
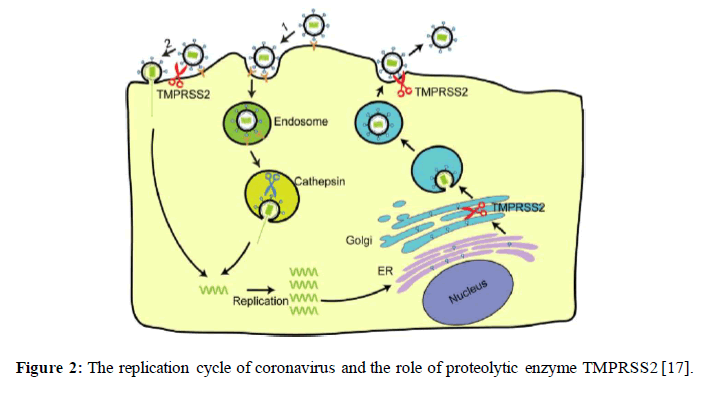
SARS-coV-2 targets human angiotensin converting enzyme-2 (ACE-2) receptor enzyme in lungs via its RBD.9 High membrane fusion activity occurs through host cell seine protease enzyme called transmembrane serine protease 2 (TMPRSS2) at two proteolytic cleavage sites, S1/S2 boundary and S2' site which is located within upstream of putative fusion peptide [17].
This step activates the glycoprotein of COVID-19 and facilitate its entry into the target cell. Once the virus enters cytosol of human host cell, +SSRNA released, then RNA transcription and replication occur. New viral RNA is directed to endoplasmic reticulum, and assembly takes place by the aid of Golgi intermediate. Then viral RNA and differentiated protein fragments assemble and form vesicles which are transported to host cell surface and released by exocytosis process to be ready for attacking new alveoli cells [17]. Figure 2.
Development of serine protease inhibitors may help in preventing the entrance of COVID-19 into alveoli cells and block viral infection without affecting on ACE-2 receptor enzyme and its vital activity in treatment of hypertension.
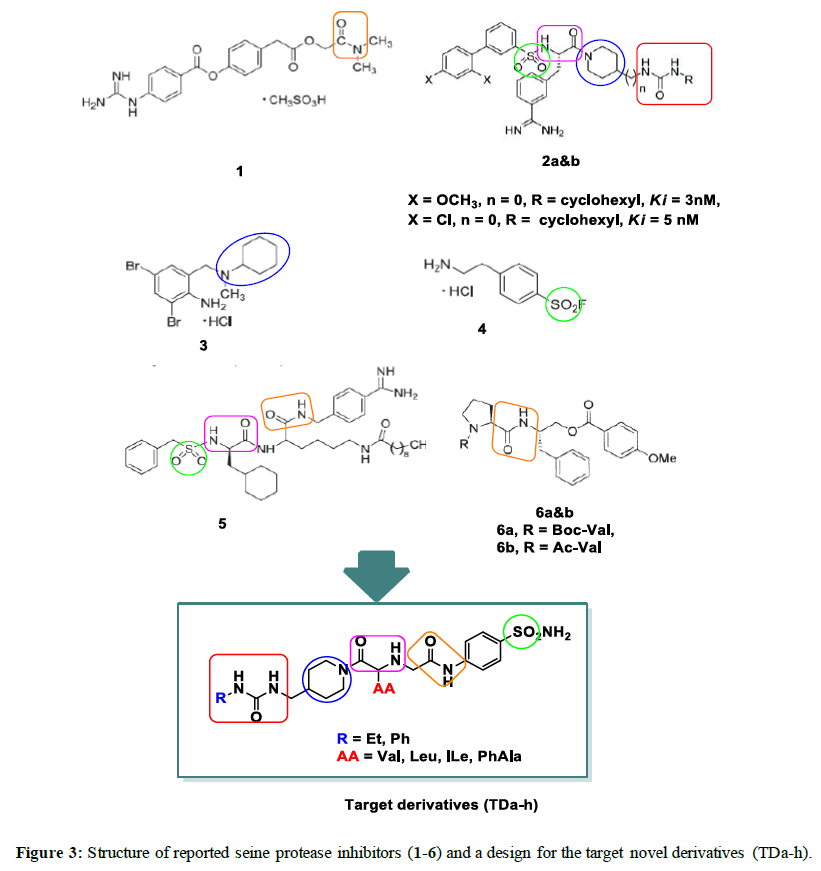
Camostat mesylate (Figure 3), a seine protease inhibitor, showed antiviral activity for SARS-coV infection, that is why we decided to design new serine protease inhibitors aiming at finding new compounds with promising activity in treatment of SARS-coV2 infection [18].
Moreover, compounds (2a&b, Figure 3), 3-amidinophenylalanine derivatives could inhibit TMPRSS2 possessing Ki values ≤ 5 nM [19]. In addition, bromohexine hydrochloride (3, Figure 3), a mucolytic cough suppressant, is also effective as TMPRSS2 inhibitor with IC5 =0.75 μM, in case of influenza virus and coronavirus infections [17].
4-(2-Aminomethyl) benzenesulfonyl fluoride hydrochloride (4, Figure 3), is one of FDA-approved serine protease inhibitors. It showed varying degrees of anti-viral activities [20].
It was also reported that benzamide –derived peptide mimetic derivative (5, Figure 3) exhibited potent TMPRSS2 inhibitory activity against H1N1 influenza virus [21].
Inhibition of serine proteases could be achieved by preparation of peptidyl inverse substrates (6a&b, Figure 3) by esterification of β-amino alcohols with p-methoxybenzoic acid. These inhibitors exhibited high specificity to certain seine protease enzymes. Thus Boc-Val and Ac-Val afforeded substrates with specific inhibitory activity to chymotrypsin serine protease enzyme [22]. Guided by the aforementioned reported compounds and from deep inspection to the important pharmacophores in the above reported compounds, we decided to design novel serine protease inhibitors containing:
1- Amide linkages,
2- Urea derivative,
3- Piperidyl as cyclic aliphatic amine,
4- Sulfonyl group,
5- Amino acid residues (valine, leucine, isoleucine and phenyl alanine)
All the designed compounds TDa-h were subjected to molecular docking study as well as drug-likeness and in-silico ADMET prediction.
Materials and Methods
Molecular docking calculations
All the designed compounds were built using ChemDraw Ulta 12.0. They were energy minimized by using MOE 2014.0901 system. The docking experiment was carried out using TMPRSS2 enzyme (PDB ID: 5CE1) linked to its ligand [23]. Downloaded enzyme and its ligand were prepared for docking by removing water molecules and incomplete residues then hydrogen added and the optimized protein was saved as pdb file, which in turn used for molecular docking studies. Ligand derivative was docked and the best pose and binding score was selected. The lead compound, camostat mesylate as well as the designed compounds was prepared for the docking experiment. The prepared data base was used for docking. The results of the docking scores and H-bonding interactions for the best poses were recorded in Table 1.
Table 1: Results of molecular docking study of designed compounds TDa-h and ligand inside serine protease active site.
| Compd. No. | Binding Energy | No. of H-bonds | Interacting amino acids |
|---|---|---|---|
| (K cal/mol) | |||
| TDa | -6.87224436 | 1 | Ala343 |
| TDb | -7.2998848 | 3 | Ala343 |
| Cys381 | |||
| Gln350 | |||
| TDc | -6.98205328 | 1 | Ala343 |
| TDd | -6.91680193 | 3 | Ala343 |
| Asn209 | |||
| Arg208 | |||
| TDe | -8.216 | 3 | Cys381 |
| Cys349 | |||
| Gln304 | |||
| TDf | -8.04319 | 2 | Gly380 |
| Tyr182 | |||
| TDg | -7.70803881 | 1 | Trp377 |
| TDh | -7.01473379 | 2 | Ala343 |
| Arg208 | |||
| Camostat | -7.5383 | 3 | Val339 |
| Arg208 | |||
| Cys188 | |||
| Ligand | -7.7973 | 4 | His203, Cys349, Gly380, Asp347 |
Drug-likeness properties
Molinspiration (2018.02 version) [24] was used to predict physicochemical properties and drug-likeness of the designed compounds. Molecular weight (MW), number of hydrogen-bond acceptor (n-ON), number of hydrogen-bond donors (n-OHNH), partition coefficient (logP), number of rotable bonds (n-rotb), topological polar surface area (TPSA) and molecular volum (MV) were calculated. Both acceptable values and predicted results were summarized in Table 2.
Table 2: Drug-likeness and physicochemical properties of compounds TDa-h.
| Compd. | MW (kDa) | n-ON | n-OHNH | LogP (o/w) | n-rotb | MV | TPSA (Å) |
|---|---|---|---|---|---|---|---|
| (g/mol) | |||||||
| Acceptable value | ≤ 500 | ≤ 10 | ≤ 5 | ≤ 5 | ≤ 10 | ˂ 500 | ˂ 160 |
| TDa | 496.62 | 7 | 5 | 0.76 | 10 | 453.04 | 162.72 |
| TDb | 510.65 | 7 | 5 | 0.91 | 11 | 469.85 | 162.72 |
| TDc | 510.65 | 7 | 5 | 0.97 | 11 | 469.85 | 162.72 |
| TDd | 544.67 | 7 | 5 | 1.25 | 11 | 491.31 | 162.72 |
| TDe | 544.67 | 7 | 5 | 1.5 | 10 | 491.09 | 162.72 |
| TDf | 558.69 | 7 | 5 | 1.91 | 11 | 507.89 | 162.72 |
| TDg | 558.69 | 7 | 5 | 1.84 | 11 | 507.89 | 162.72 |
| TDh | 592.71 | 7 | 5 | 2.02 | 11 | 529.35 | 162.72 |
In silico ADME properties
In silico ADME properties were calculated using PreADME online surver. Human intestinal absorption (HIA), cell permeability of CaCo-2 cell, skin permeability (SP), blood brain barrier (BBB) and plasma protein binding (PPB) were calculated and predicted results were listed in Table 3.
Table 3: Predicted in silico ADME properties for target compounds TDa-h.
| Compd. | Absorption | Distribution | Excretion | ||
|---|---|---|---|---|---|
| No. | aHIA | bCaCo-2 | cSkinP | dBBB | ePPB |
| (logKp) | |||||
| Acceptable value | 70-100% | ˃ 70 | ˃ 90 | ˃ 0.40 | ˃ 90% |
| (good absorption) | (high permeability) | (high permeability) | (CNS active compound) | ||
| TDa | 71.13 | 0.63 | -2.84 | 0.03 | 20.87 |
| TDb | 73.6 | 0.64 | -2.74 | 0.03 | 26.34 |
| TDc | 73.6 | 0.64 | -2.73 | 0.03 | 26.52 |
| TDd | 84.39 | 0.7 | -2.5 | 0.04 | 31.68 |
| TDe | 84.51 | 0.81 | -2.47 | 0.04 | 39.05 |
| TDf | 85.22 | 0.81 | -2.41 | 0.04 | 45.02 |
| TDg | 85.22 | 0.82 | -2.41 | 0.04 | 47.35 |
| TDh | 88.36 | 0.88 | -2.3 | 0.04 | 64.13 |
aHuman intestinal absorbtion (%), bin vitro CaCo cell permeability (nm/sec), cSkin permeability, din-vitro blood brain barrier penetration (C. brain/C. blood), ein-vitro plasma protein binding (%).
Metabolic prediction
Metabolism prediction for the designed compounds was examined using online preADMET server. The most important metabolic parameter tested was cytochrome P450 (CYP) isoforms, CYP2C19, CYP2C9, CYP2D6 and CYP3A4.
Toxicity prediction
AMES test, rodent carcinogenicity assay and hERG-inhibition were examined to predict toxicity properties for the designed compounds by using preADMET online server [24].
Conclusion
Searching for effective therapeutic agents for treatment of COVID-19 is considered as a major challenge that faces scientific researchers all over the world. Taking into consideration that camostat mesylate is the most acceptable and FDA approved drug as serine protease inhibitor, so designing of new agents mimic it in their chemical structure may be a privilege stone for finding therapeutic agents that help in treatment of this pandemic disease.
All the designed compounds were subjected to molecular docking experiment inside TMPRSS2 active site, four derivatives TDd, TDe, TDf and TDh shared the same amino acids observed from docking of ligand and/or camostat derivatives.
Using compoutational analysis, it eas observed that compound TDa, exhibited good drug-likeness properties (Lipiniski's rule). Most of the compounds showed in silico ADMET predicted values. Hence, the designed compounds could be a promising drug leads for future development of anti-SARS-Cov-2 drugs aiming to stop coronaviruses spread.
Conflict of interest
The authors have no conflict of interest to declare.
References
[1] A.R. Fehr, R. Channappanavar and S. Perlman, Annual Review of Medicine. 2017. 68: p. 387.
[2] J.S.M. Peiris, C.M. Chu, V.C.C. Cheng et al., Lancet. 2003. 361: p. 1767.
[3] R.J. de Groot, S.C. Baker, R.S. Baric et al., J. Vir. 2013. 87: p. 7790.
[4] M. Hoffmann, H. Kleine-Weber, S. Schroeder et al., Cell, 2020. 181(2): p. 271.
[5] C.L. Huang, Y.M. Wang, X.W. Li et al., China. Lancet. 2020. 395(10223): p. 497.
[6] C. Wang, P.W. Horby, F.G. Hayden et al., Lancet, 2020. 395(10223): p. 470.
[7] P. Zhou, X.L. Yang, X.G. Wang et al., Nature, 2020. 579 (7798): p. 270.
[8] E. de Wit, N. van Doremalen, D. Falzarano et al., Nature Reviews Microbiology. 2016. 14(8): p. 523.
[9] D. Bestle, M.R. Heindl, H. Limburg et al., 2020.
[10] M. Hoffmann, H. Kleine-Weber, N. Krüger et al., 2020.
[11] D. Wrapp, N.S. Wang, K.S. Corbett et al., Science, 2020. 367(6483): p. 1260.
[12] A.C. Walls, Y.J. Park, M.A. Tortorici et al., Cell, 2020. 181(2): p. 281.
[13] Y.S. Wan, J. Shang, R. Graham et al., J. Vir. 2020. 94 (7).
[14] W.H. Li, M.J. Moore, N. Vasilieva et al., Nature, 2003. 426 (6965): p. 450.
[15] J.K. Millet and G.R.Whittaker, Virus Research, 2015. 202: p. 120.
[16] J. Reguera, G. Mudgal, C. Santiago et al., Virus Research, 2014. 194: p. 3.
[17] L.W. Shen, H.J. Mao, Y.L. Wu et al., Biochimie, 2017. 142: p. 1.
[18] Y. Zhou, P. Vedantham, K. Lu et al., Antiviral Research, 2015. 116: p. 76.
[19] J. Sturzebecher, D. Prasa, J. Hauptmann et al., J. Med. Che. 1997. 40 (19): p. 3091.
[20] M. Yamamoto, S. Matsuyama, X. Li et al, Antimicrobial Agents and Chemotherapy, 2016. 60 (11): p. 6532.
[21] E. Bottcher-Friebertshauser, C. Freuer, F. Sielaff et al., J. Vir, 2010. 84 (11): p. 5605.
[22] B. Walker and J.F. Lynas, Cellular and Molecular Life Sciences, 2001. 58 (4): p. 596.
[23] https://www.rcsb.org/structure/5CE1.
[24] J.C. Cole, J.W.M. Nissink and R.Taylor. Virtual screening in drug discovery. 2005.