Research Article - Der Pharma Chemica ( 2018) Volume 10, Issue 10
The In-Silico Screening of Pyrazolo[3,4-E][1,4]Thiazepine Derivatives as Potent Cytotoxic Agents
Srikanth A1, Hemanth Kumar P1, Vijayakumar V1, Sudha2, Anand A1,2 and Sarveswari S1*
1Centre for Organic and Medicinal Chemistry, VIT University, Vellore 632 014, India
2Medical and Biological Computing Laboratory, School of Biosciences and Technology, VIT University, Vellore 632 014, India
- *Corresponding Author:
- Sarveswari S
Centre for Organic and Medicinal Chemistry
VIT University
Vellore 632 014, India
Abstract
A new series of 1, 4-thiazepinone incorporated pyrazolopyridines has been synthesized using L-proline as a catalyst through multi-component reaction which in turn subjected to the in-silico cytotoxicity studies against various cancer cell lines and almost all the compounds found to show good to moderate activity. Among the tested compounds, compound 4f and 4k are found to have le
Keywords
1,4-thiazepine derivatives, L-proline catalysis, C-S bond formation, Docking studies.
Introduction
Design and synthesis of new lead structures essentially interacting with novel biological targets are the major challenges for medicinal chemists and biologists. It is necessary to design target oriented synthesis for the development of new molecules even though a large number of drugs are in clinical trials [1-3]. There is a considerable research over design of new drugs which are more selective for cancer cells with lesser side effects. Chemotherapy in cancer treatment is utilizing the intracellular barrier crossing property of drugs [4,5].
3,4-di hydro-5-oxo-1,4-benzothiazepine alkyl derivatives were described as HIV-1 enzyme integrase [6], reverse transcriptase inhibitors [7], and antitumor agents [8]. The core unit hetero ring systems are potent inhibitors of HSV-1 replication [9], compounds possessing H1 antihistamine activity [10], selective antagonists of 5-HT1A and dopamine D2 receptors [11]. Substrate organization, transition state stabilization and product release are the characteristic features of enzymes and catalysts, where convergent construction of molecular frame works for complex skeletons, eco-friendliness and chemo selectiveness are additive features.
Materials and Methods
We attempted an L-proline catalyzed synthesis of 1, 4-thiazepinones (Scheme 1) and screened them for their cytotoxic activity [12] based on target binding ability observed in in-silico studies (Table 1). The formation of C-S bond is a common functionality found in various pharmaceutically active and bio molecules [13,14] as the viscous mass.
| Compound | ArCHO (3a-m) | Time (Hour) | Yield (%)b | MP (°C) |
|---|---|---|---|---|
| 4a | 2-Methoxybenzaldehyde | 2 | 92 | 123-125 |
| 4b | 3, 4, 5-Trimethoxybenzaldehyde | 2 | 89 | 115-117 |
| 4c | 3-Nitrobenzaldehyde | 2.45 | 76 | 130 |
| 4d | 2,5-Dimethoxybenzaldehyde | 2 | 90 | 118-120 |
| 4e | 2,4-Dichlorobenzaldehyde | 3 | 79 | 238-240 |
| 4f | 4-Cyanobenzaldehyde | 2.3 | 72 | 196-198 |
| 4g | 2-Nitrobenzaldehyde | 2.3 | 75 | 132-136 |
| 4h | 3,4-Dimethoxybenzaldehyde | 2.15 | 88 | 218-220 |
| 4i | 2, 4, 6-trimethoxybenzaldhyde | 2.3 | 91 | 110-112 |
| 4j | 4-Chlorobenzaldehyde | 2 | 86 | 120 |
| 4k | 2-Fluorobenzaldehyde | 3 | 82 | 138-141 |
| 4l | 4-Methylbenzaldehyde | 2 | 89 | 190-192 |
| 4m | 4-Methoxybenzaldehyde | 2.15 | 87 | 180-182 |
Table 1: Optimization of 1,4-thiazepine fused pyrazolines (4a-m) in ethanol solvent medium
Results and Discussions
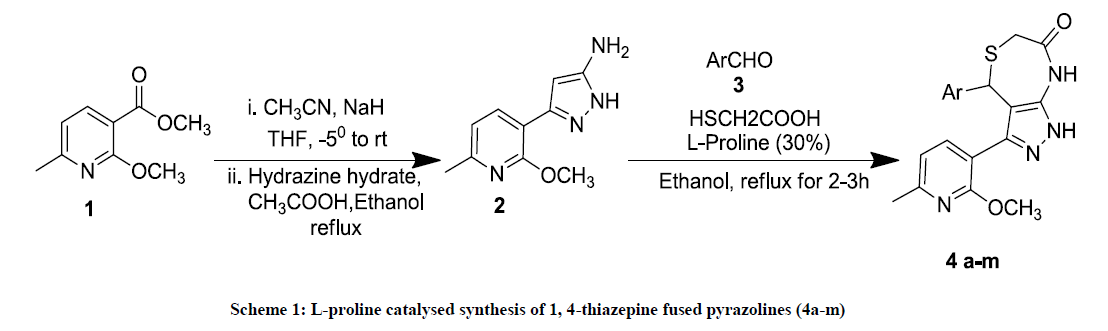
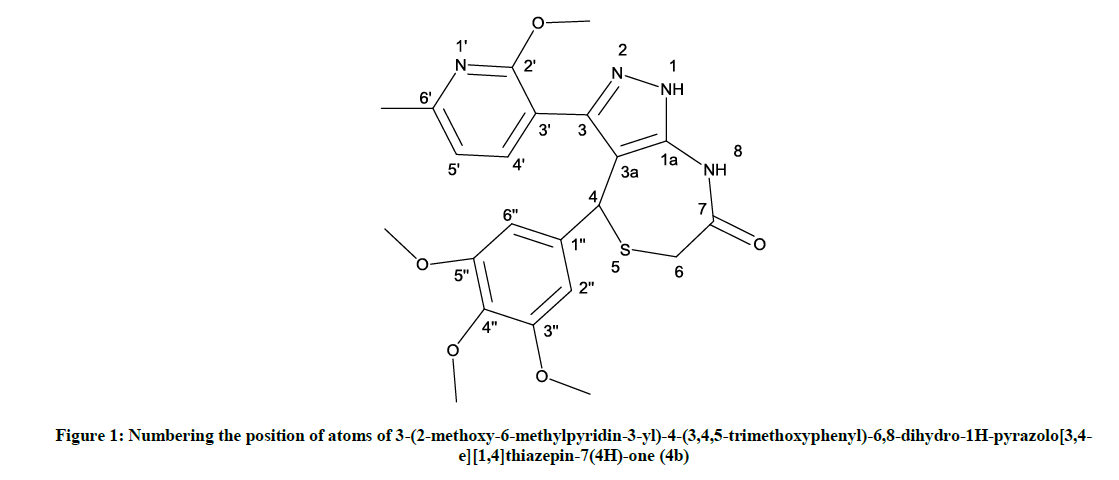
The newly synthesized 1, 4-thiazepines 4a-m was characterized [12] by Proton Nuclear Magnetic Resonance (1H-NMR), Carbon-13 Nuclear Magnetic Resonance (13C-NMR) and HRMS (Figure 1).
In silico studies of Pyrazolo pyridine fused 1,4-thiazepines (4a-m)
Newly synthesized compounds 4a-m has been subjected to the in-silico studies towards six selected target proteins. The 3-dimension (3D) coordinates of six cancer target proteins were selected and obtained from protein data bank (PDB) [15]. The PDB id 3B8Q (renal cancer) and 4FLH (colon cancer), 1SVC (pancreatic cancer), 1MOX (lung cancer), 2DSQ (normal breast epithelium cancer) and 3TWJ (non-small cell lung cancer) were selected for structural analysis based on its high resolution crystallographic structure. The co-crystallised ligands and water molecules were identified and removed from the atomic coordinate file and the structure was optimized and refined using spdbv viewer [16]. The synthesized compounds structure was sketched with the help of chemsketch [17]. 3-dimensional conversion and geometry optimization of all the compounds were performed using chimera for flexible conformations of the compounds during the docking [18]. Data are given in Tables 2-7. iGEMDOCK2.1(A generic evolutionary method for molecular docking) docking software was used to study the detailed intermolecular interactions between the target protein and the ligand molecule [19]. iGEMDOCK integrated the molecular docking, post screening analysis and visualization steps. We selected Rho-associated protein kinase 1, (PDB Id: 3TWJ), nuclear factor kappa-b (nf-kb) (PDB Id: 1SVC), vascular endothelial growth factor receptotor-2 (PDB Id: 3B8Q) and human phosphoinositide 3-kinase (PI3K-gamma) (PDB Id: 4FLH), crystal structure of human epidermal growth factor receptor (PDB id: 1MOX) and crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor (PDB Id: 3CE3) as target proteins to carry out the docking analysis of our synthesized compounds. The 3D atomic coordinates of each therapeutic target protein were implemented through the GEMDOCK graphical environment interface. Then the default option was used to import the 3D coordinates of 13 synthesized compounds. Before docking the output path was set. GEMDOCK default parameters included the population size (n=200), generation (g=70) and number of solutions (s=10) to compute the possible binding conformation of synthesized compounds. Then the docking run was started using Gemdock scoring function. After docking, the individual binding conformation of each ligand was observed and their binding affinity with the target proteins was analyzed. The best binding pose and binding energy of each ligand was selected. In the post docking analysis, Vander Waals score, Z-score and the details of interacted residues were saved in output folder. Protein-ligand binding site was analysed and visualized using pymol [20].
| Sample Code | Z Score E (pharma) | Total Energy | VDW |
|---|---|---|---|
| 4a | -132.3 | -106.1 | -94.9414 |
| 4b | -104.8 | -104.8 | -80.6809 |
| 4c | -107.9 | -107.9 | -81.1659 |
| 4d | -139.6 | -105.4 | -110.049 |
| 4e | -100.4 | -100.4 | -69.5653 |
| 4f | -159.5 | -120.5 | -115.7825 |
| 4g | -110.2 | -115.3 | -75.694 |
| 4h | -111 | -111 | -81.1596 |
| 4i | -107.9 | -109.9 | -108.515 |
| 4j | -136.4 | -108.5 | -101.622 |
| 4k | -120 | -116.5 | -88.9061 |
| 4l | -103.5 | -104 | -82.4252 |
| 4m | -100.4 | -102.2 | -76.556 |
Table 2: Docking results of synthesized compounds in the binding site nuclear factor kappa-b (pancreatic cancer)
| Sample Code | Z Score E(pharma) | Total Energy | VDW |
|---|---|---|---|
| 4a | -117.7 | -96.8184 | -82.7337 |
| 4b | -135.1 | -107.591 | -96.591 |
| 4c | -104.4 | -101.006 | -77.4242 |
| 4d | -105.3 | -103.284 | -77.612 |
| 4e | -138.8 | -109.491 | -109.491 |
| 4f | -150.6 | -124.516 | -118.3808 |
| 4g | -138.3 | -107.206 | -100.556 |
| 4h | -140 | -110.913 | -107.413 |
| 4i | -141.7 | -107.642 | -97.5849 |
| 4j | -101.1 | -97.7 | -106.654 |
| 4k | -148.2 | -118.243 | -102.321 |
| 4l | -107.4 | -105.821 | -98.458 |
| 4m | -130.2 | -108.287 | -101.347 |
Table 3: Docking results of synthesized compounds in the binding site vascular endothelial growth factor receptor 2 (Renal cancer)
| Sample code | Z Score E(pharma) | Total Energy | VDW |
|---|---|---|---|
| 4a | -171.5 | -113.767 | -85.8677 |
| 4b | -128.7 | -117.116 | -101.019 |
| 4c | -122.2 | -114.124 | -82.8858 |
| 4d | -146.2 | -108.713 | -88.4499 |
| 4e | -146.2 | -122.03 | -101.328 |
| 4f | -176.3 | -130.02 | -101.1308 |
| 4g | -176.8 | -109.369 | -70.0512 |
| 4h | -156.8 | -117.387 | -70.187 |
| 4i | -134.1 | -123.515 | -90.7239 |
| 4j | -156.6 | -113.767 | -85.8677 |
| 4k | -176.6 | -124.116 | -113.019 |
| 4l | -109.4 | -120.211 | -100.337 |
| 4m | -147.5 | -108.713 | -88.4499 |
Table 4: Docking results of synthesized compounds in the binding site human phosphoinositide-3-kinase (pi3k)-gamma (colon cancer)
| Sample code | Z Score E (pharma) | Total Energy | VDW |
|---|---|---|---|
| 4a | -132.4 | -101.064 | -83.905 |
| 4b | -104.2 | -110.164 | -89.7346 |
| 4c | -111.8 | -109.114 | -75.5307 |
| 4d | -127.4 | -99.2605 | -83.5389 |
| 4e | -106.6 | -104.302 | -84.7897 |
| 4f | -112.2 | -119.094 | -89.0001 |
| 4g | -110.4 | -110.386 | -67.4132 |
| 4h | -109.6 | -108.446 | -79.125 |
| 4i | -147.1 | -102.852 | -90.1644 |
| 4j | -125.6 | -109.055 | -89.5523 |
| 4k | -130.7 | -85.516 | -97.7281 |
| 4l | -115 | -105.664 | -83.4292 |
| 4m | -132.4 | -108.867 | -91.9362 |
Table 5: Docking results of synthesized compounds in the binding site Hepatocyte growth factor receptor (Non-small cell lung cancer)
| Sample code | Z Score E (pharma) | Total Energy | VDW |
|---|---|---|---|
| 4a | -175.9 | -124.5 | -93.7778 |
| 4b | -121.7 | -121.7 | -103.5 |
| 4c | -122.6 | -120.6 | -78.7755 |
| 4d | -113.9 | -113.9 | -94.5327 |
| 4e | -152.4 | -105.5 | -84.8099 |
| 4f | -136.3 | -127.3 | -81.0741 |
| 4g | -166.8 | -115.5 | -88.6983 |
| 4h | -157.9 | -108.4 | -83.0514 |
| 4i | -112.3 | -112.3 | -74.7006 |
| 4j | -105.8 | -105.8 | -88.7077 |
| 4k | -160.1 | -95.1 | -98.4659 |
| 4l | -164.7 | -117.9 | -87.9991 |
| 4m | -136.5 | -102.1 | -81.0066 |
Table 6: Docking results of synthesized compounds in the binding site of epidermal growth factor receptor (human epidermal growth factor receptor; lung cancer)
| Sample code | Z Score E (pharma) | Total Energy | VDW |
|---|---|---|---|
| 4a | -173.6 | -101.931 | -98.0808 |
| 4b | -181 | -90.642 | -103.906 |
| 4c | -183.3 | -97.482 | -92.023 |
| 4d | -180.8 | -100.698 | -103.063 |
| 4e | -169.7 | -94.953 | -88.1788 |
| 4f | -183.5 | -105.169 | -102.391 |
| 4g | -119.7 | -101.374 | -92.0233 |
| 4h | -171.7 | -92.479 | -86.6902 |
| 4i | -159.2 | -103.653 | -89.329 |
| 4j | -112 | -96.03 | -91.1223 |
| 4k | -120.8 | -80.364 | -80.9456 |
| 4l | -156 | -102.064 | -95.9908 |
| 4m | -103 | -101.509 | -96.8364 |
Table 7: Docking results of synthesized compounds in the binding site of Rho-associated protein kinase (Normal breast epithelium cancer)
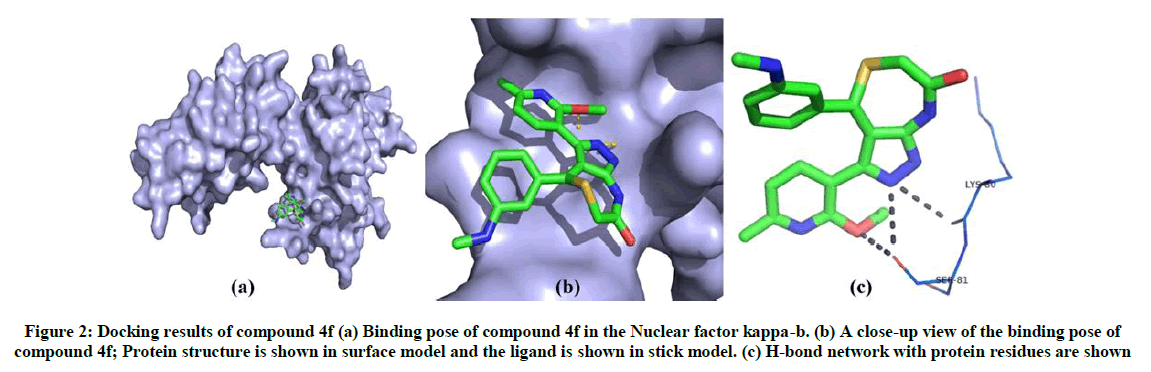
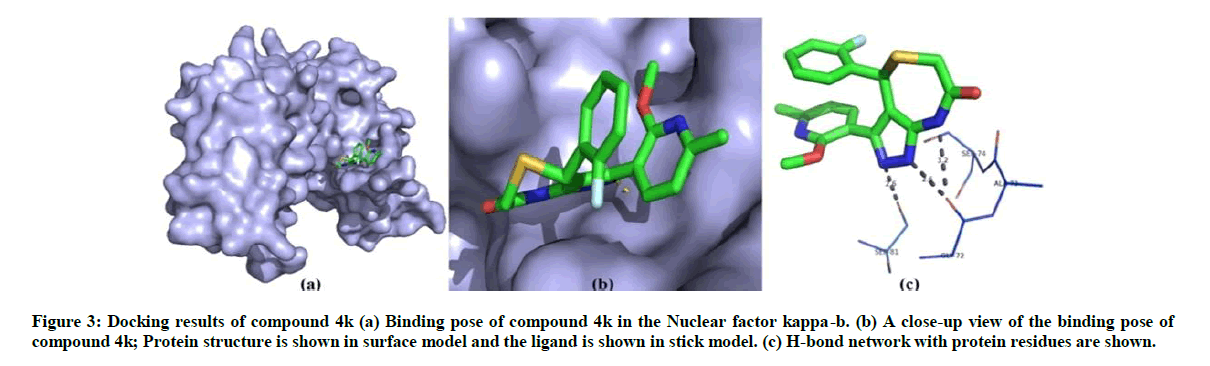
The binding affinity of 13 synthesised compounds towards nuclear factor kappa-b has been investigated and compounds 4f and 4k found to exhibit best binding conformation with nuclear factor kappa-b. The binding energy and Z-score values of compound 4f and 4k are given in Table 2. On analysis of the interaction and position of compounds 4f and 4k in the nuclear factor kappa-b binding site, it is observed that compound 4f forms three H-bonds with nuclear factor kappa-b and compound 4k shows three hydrogen bond interaction with the amino acid residues such as Ser 81, Ser 74 and Gly 72. The binding pose and interaction mode of compound 4f and 4k are shown in Figures 2 and 3.
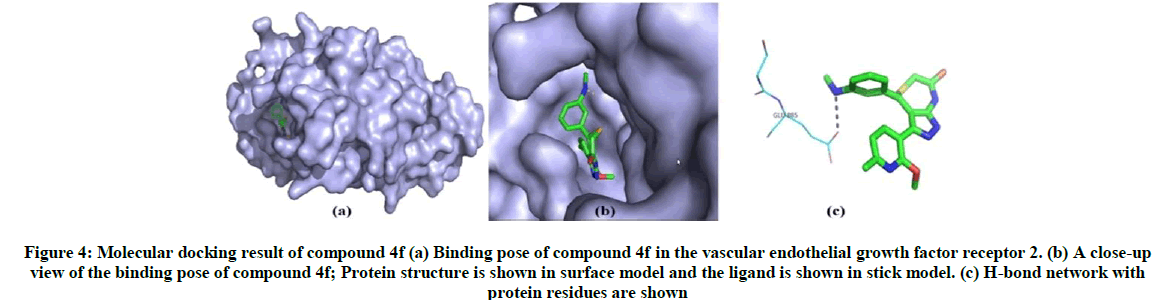
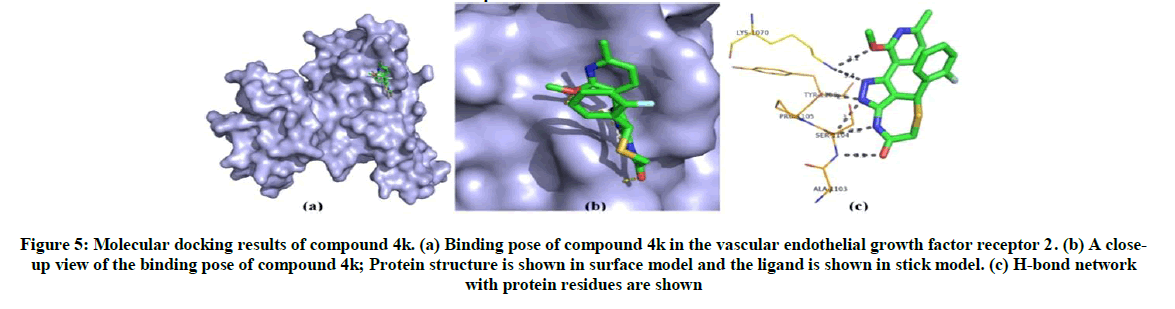
The binding energy is calculated for 13 synthesized ligand molecules with vascular endothelial growth factor receptor 2 which has play important role renal cancer. From the docking results, compound 4f and compound 4k has shown best conformation with vascular endothelial growth factor receptor 2. The binding energy and Z-score value of compound 4f and compound 4k is given in Table 3. Compound 4f binds to binding site of vascular endothelial growth factor receptor 2. It can be seen in Figure 4 and the nitrogen atoms of compound 4f forms only one hydrogen bond with Glu 885. On analysis of the interaction and position of compound 4k in thevascular endothelial growth factor receptor 2 binding site, it is observed that compound 4k forms six H-bonds with Lys1070, Tyr1106, Pro1105, Ser1104 and Ala1103. The best possible binding mode of compound 4f and 4k with vascular endothelial growth factor receptor 2 corresponding 2D interaction models are depicted in Figures 4 and 5.
Figure 4: Molecular docking result of compound 4f (a) Binding pose of compound 4f in the vascular endothelial growth factor receptor 2. (b) A close-up view of the binding pose of compound 4f; Protein structure is shown in surface model and the ligand is shown in stick model. (c) H-bond network with protein residues are shown
Figure 5: Molecular docking results of compound 4k. (a) Binding pose of compound 4k in the vascular endothelial growth factor receptor 2. (b) A close-up view of the binding pose of compound 4k; Protein structure is shown in surface model and the ligand is shown in stick model. (c) H-bond network with protein residues are shown
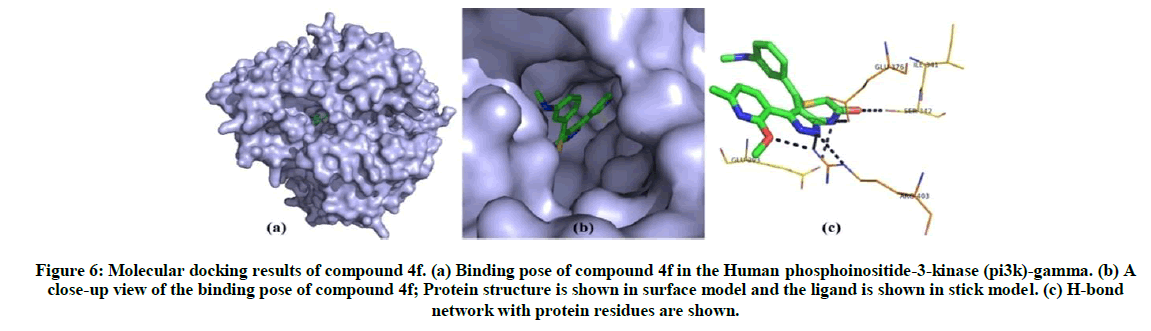
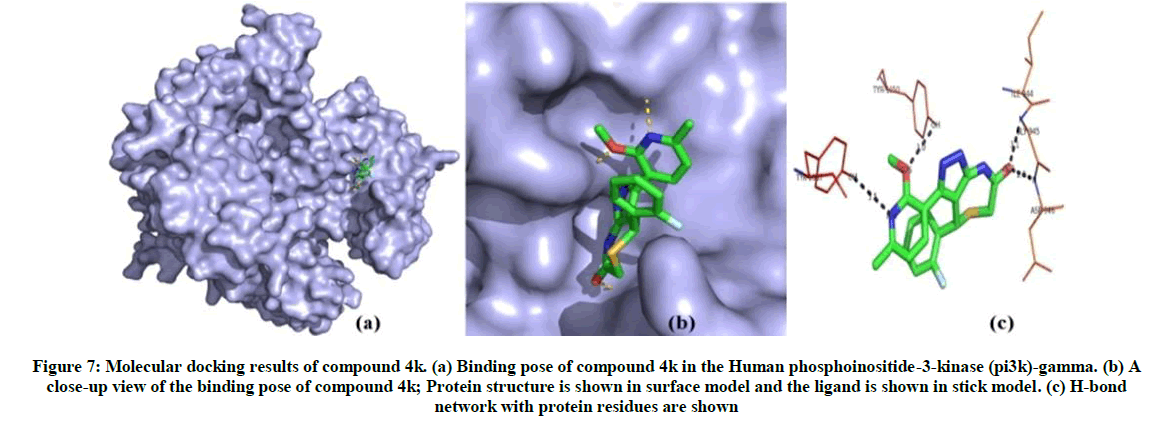
The binding energy is calculated by iGEMDOCK for synthesised compounds. Compound 4f and 4k has shown best conformation with human phosphoinositide-3-kinase involved in colon cancer. The binding energy and Z-score value of the compound 4f and 4k is given in Table 4. Compound 4f binds to the binding site and forms five hydrogen bonds with human phosphoinositide-3-kinase. Glu176, Ser342, Arg403, Ile341 and Glu293 are involved in the interaction. From the analysis of interaction and position of compound 4k in the human phosphoinositide-3-kinase, it is observed that four hydrogen bonds are found and the amino acid residues Tyr1050, Asp946, Gly945 and Ile944 and Tyr1067 are participated in the interaction. The hydrogen bond network and binding pose of compound 4f and compound 4k are shown in Figures 6 and 7.
Figure 6: Molecular docking results of compound 4f. (a) Binding pose of compound 4f in the Human phosphoinositide-3-kinase (pi3k)-gamma. (b) A close-up view of the binding pose of compound 4f; Protein structure is shown in surface model and the ligand is shown in stick model. (c) H-bond network with protein residues are shown.
Figure 7: Molecular docking results of compound 4k. (a) Binding pose of compound 4k in the Human phosphoinositide-3-kinase (pi3k)-gamma. (b) A close-up view of the binding pose of compound 4k; Protein structure is shown in surface model and the ligand is shown in stick model. (c) H-bond network with protein residues are shown
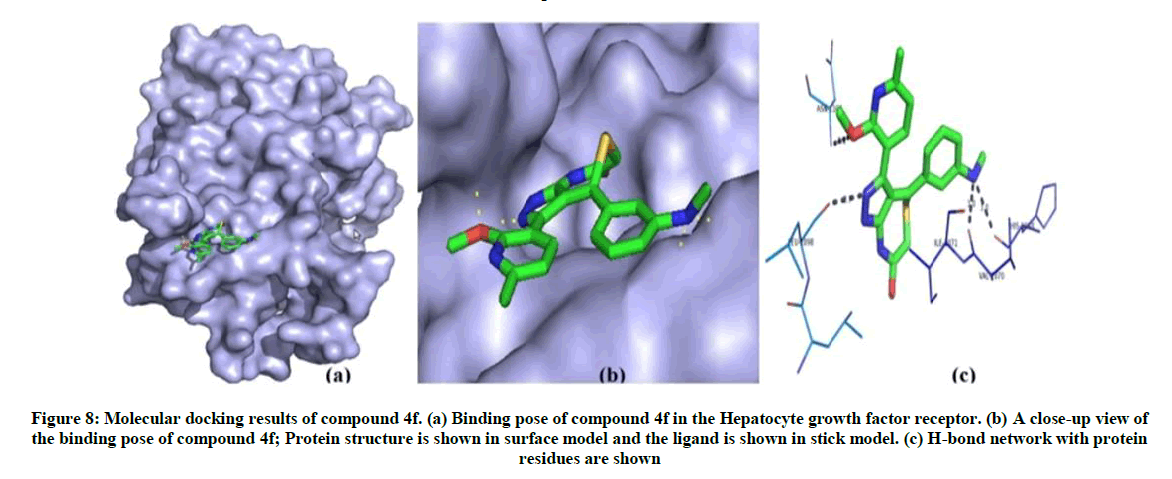
Docking analysis of compound 4f shows best binding conformation with hepatocyte growth factor receptor which has crucial role in non-small cell lung cancer (energy=-119.094 K. cal/Mol, Z-score=-112.2) (Table 5). From the Figure 8, it has shown that the compound 4f forms four hydrogen bonds with hepatocyte growth factor receptor. The binding pose and hydrogen bond network of compound 4f in hepatocyte growth factor receptor are shown in Figure 8.
Figure 8: Molecular docking results of compound 4f. (a) Binding pose of compound 4f in the Hepatocyte growth factor receptor. (b) A close-up view of the binding pose of compound 4f; Protein structure is shown in surface model and the ligand is shown in stick model. (c) H-bond network with protein residues are shown
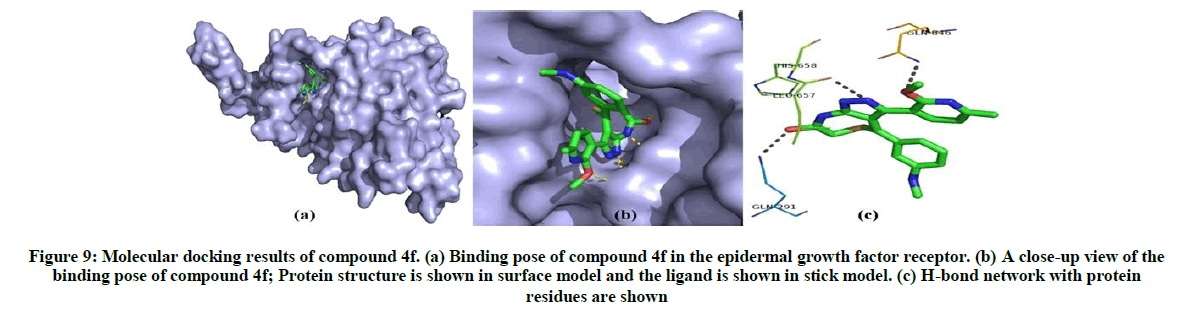
The post docking analysis of compound 4f shows best binding pose in epidermal growth factor receptor which has key role in lung cancer development (Total energy=-127.3 Kcal/Mol, Z-score=-136.3) (Table 6). Compound 4f binds to epidermal growth factor receptor and form three H-bond interactions with His658, Gln846 and Gln291 residues. The best binding pose of compound 4f in the epidermal growth factor receptor and corresponding 2D interaction models, hydrogen bonds and bond distance are depicted in Figure 9.
Figure 9: Molecular docking results of compound 4f. (a) Binding pose of compound 4f in the epidermal growth factor receptor. (b) A close-up view of the binding pose of compound 4f; Protein structure is shown in surface model and the ligand is shown in stick model. (c) H-bond network with protein residues are shown
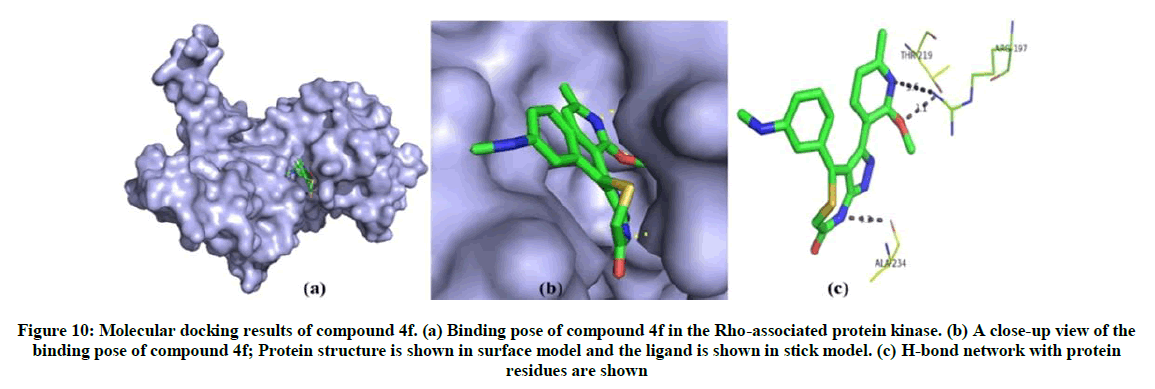
From the docking analysis of synthesised compounds and Rho-associated protein kinase, compound 4f has high binding affinity (energy=-105.169 Kcal/Mol, Z-score=-183.5) and shows three hydrogen bond interactions with the residues Thr219, Arg197 and Ala234. The docking results of all 13 ligand molecules with Rho-associated protein kinase are given in Table 7. The possible binding mode of compound 4f in the Rho-associated protein kinase binding site and corresponding 2D interaction models, hydrogen bonds are shown in Figure 10.
Figure 10: Molecular docking results of compound 4f. (a) Binding pose of compound 4f in the Rho-associated protein kinase. (b) A close-up view of the binding pose of compound 4f; Protein structure is shown in surface model and the ligand is shown in stick model. (c) H-bond network with protein residues are shown
Among the 13 compounds, compound 4f and 4k are found to have least binding energy value and Z-score value. These compounds found to form more stable ligand-receptor complex amongst other compounds. Compound 4f shows best binding pose with all six selected cancer target proteins and compound 4k shows best binding pose with nuclear factor kappa-b (pancreatic cancer), vascular endothelial growth factor receptor 2 (renal cancer) and human phosphoinositide-3-kinase (colon cancer). In examining the interaction of compound 4f with all six cancer target proteins, it is observed that compound 4f shows more than three hydrogen bond interactions with the target proteins except vascular endothelial growth factor receptor 2.
Conclusion
In summary a series of pyrazolopyridine fused 1, 4-thiazepinones has been synthesized using MCR with L-proline as a catalyst, which in turn subjected to the in-silico studies against various cancer cell lines and almost all the compounds found to possess good to moderate activity. Among these, the compounds 4f and 4k found to possess activity equal to the control in all tested cell lines docking analysis.
Acknowledgement
The authors are thankful to VIT University Vellore, India for providing facilities to carry out research work, and also thankful to SAIF, Vellore Institute of Technology, SAIF, IIT-Madras and VIT-TBI for providing NMR, Mass and IR spectral facilities respectively.
References
- M.D. Burke, S.L. Schreiber, Angew. Chem. Int. Ed., 2004, 43, 46.
- S. Oh, H.J. Jang, S.K. Ko, Y. Ko, S.B. Park, J. Comb. Chem. 2010, 12, 548.
- S.L. Schreiber, Science., 2000, 287, 1964.
- S.P. Gupta, Chem. Rev., 1994, 94, 1507.
- P. Martins, J. Jesus, S. Santos, L.R. Raposo, C.R. Rodrigues, P.V. Baptista, A.R. Fernandes, Molecules., 2015, 20, 16852.
- N. Neamati, J.A. Turpin, H.E. Winslow, J.L. Christensen, K. Williamson, A. Orr, W.G. Rice, Y. Pommier, A. Garofalo, A. Brizzi, G. Campiani, I. Fiorini, V. Nacci, J. Med. Chem., 1999, 42, 3334.
- H. Maruenda, F. Johnson, J. Med. Chem., 1995, 38, 2145.
- A. Garofalo, G. Campiani, I. Fiorini, V. Nacci, Farmaco., 1993, 48, 275.
- S.L. Boulware, J.C. Bronstein, E.C. Nordby, P.C. Weber, Antivir. Res., 2001, 51, 111.
- A.D. Cale, T.W. Gero, K.R. Walker, Y.S. Lo, W.J. Welstead, L.W. Jaques, A.F. Johnson, C.A. Leonard, J.C. Nolan, D.N. Johnson, J. Med. Chem., 1989, 32, 2178.
- T. Nakao, H. Tanaka, H. Yamato, T. Akagi, S. Takehara, Patent 5, 141, 930, 1992, 1.
- A. Srikanth, S. Sarveswari, V. Vijayakumar, P. Gridharan, S. Karthikeyan, Med. Chem. Res., 2014, 24, 553.
- L. Liu, J.E. Stelmach, S.R. Natarajan, M. H Chen, S.B. Singh, C.D. Schwartz, C.E. Fitzgerald S.J. O'Keefe, D.M. Zaller, D.M. Schmatz, J.B. Doherty, Bioorg. Med. Chem. Lett., 2003, 13, 3979.
- S.W. Kaldor, V.J. Kalish, J.F. Davies, B.V. Shetty, J.E. Fritz, K. Appelt, J.A. Burgess, K.M. Campanale, N.Y. Chirgadze, D.K. Clawson, B.A. Dressman, S.D. Hatch, D.A. Khalil, M.B. Kosa, P.P. Lubbehusen, M.A. Muesing, A.K. Patick, S.H. Reich, K.S. Su, J.H. Tatlock, J. Med. Chem., 1997, 40, 3979.
- H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne, Nucleic Acids Res., 2000, 28, 235.
- N. Guex, M.C. Peitsch, Electrophoresis., 1997, 18, 2714.
- Z. Li, H. Wan, Y. Shi, P. Ouyang, J. Chem. Inf. Comput. Sci., 2004, 44, 1886.
- E.F. Pettersen, T.D. Goddard, C.C. Huang, G.S. Couch, D.M. Greenblatt, E.C. Meng, T.E. Ferrin, J. Comput. Chem., 2004, 25, 1605.
- J.M. Yang, C.C. Chen, Proteins., 2004, 55, 288.
- D. Seeliger, B.L. de Groot, J. Comput. Aided Mol. Des., 2010, 24, 417.