Abstract
In-Silico Prediction of T- cell epitopes in VacA: First Step towards the Design of a Helicobacter Pylori Vaccine
Author(s): Elijah Kolawole Oladipo*, Abisola Temiloluwa Aina, Oluwaseyi Febenna Eko, Olubukola Monisola Oyawoye, Boluwatife Ayobami Irewolede and Temitope Isaac AdelusiHelicobacter pylori are indicated in gastritis, gastric cancer, Peptic ulcer and B cell mucosa lymphoid tissue lymphoma. The bacteria have a 50% pandemic, a higher prevalence of 75% in developing countries and 40% in developed countries. A combination of antibiotics is used to treat Helicobacter pylori infections. Some of these are Amoxicillin, tetracycline, and fluoroquinolones, but are ineffective in eliminating the infections and are also compromised by antibiotic-resistant strains, therefore a vaccine to help prevent, control and eliminate Helicobacter pylori infections is needed. We proposed to predict T-cell epitopes of VacA. Seven VacA protein sequences were retrieved from the NCBI protein database and subjected to antigenicity test using the ANTIGENpro database. All the seven (7) sequences with antigenicity score ≥0.8 were submitted to the NetCTL server and IEDB (Immune Epitope Database) MHC-II server to predict the cytotoxic T-lymphocyte (CTL) and to predict the Helper T-lymphocyte (HTL) epitopes, respectively. The cytotoxic T-lymphocyte epitopes and Helper T-lymphocyte epitopes were predicted. The seven amino acid sequences obtained from NCBI subjected to antigenicity test passed by having a score of ≥0.8. Ninety-eight (98) CTL (9-mer) ligands in total were predicted for the seven selected proteins submitted to NetCTL sever. Fifty-six (56) epitopes were selected according to their high scores. Ninety-nine (99) (15-mer) total HTL epitopes were predicted. Their IC50 value was not greater than 500nm with percentile rank ranging from 0.030-4.00, while ninety-four (94) epitopes were selected. A total of fifty-six (56) CTL and Ninety-four (94) HTL epitopes were merged using AAY, EAAK, and GPGPG linkers with sequence APPHALS as an adjuvant to construct the multi-epitope vaccine. The predicted peptides are presumed to be valuable in the multi-epitope vaccines, non-allergen without compromising the population coverage. VacA has potential B-cell and T-cell epitopes distributed throughout the sequence. Thus several fragments were identified as valuable candidates for subunit vaccines against Helicobacter pylori.
Select your language of interest to view the total content in your interested language
Google Scholar citation report
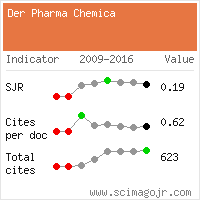
Citations : 15261
Der Pharma Chemica received 15261 citations as per Google Scholar report
Der Pharma Chemica peer review process verified at publons