Abstract
Three dimensional quantitative structure analysis substituted 1,3-diaryl propenone derivatives as antimalarial activity
Author(s): Rajesh Sharma and Swaraj PatilMolecular modeling analysis performed by k nearest neighbor molecular field analysis (kNN MFA) to recognize the necessary structural requirements of 1,3-diaryl propenone derivatives in 3D chemical space for adjusting modulation of the antimalarial activity. In study 14 compounds were selected randomly, using sphere exclusion (SE) algorithm and random selection method struture divided into training and test set. kNN-MFA methodology with stepwise (SW), simulated annealing (SA) and genetic algorithm (GA) was used for building the QSAR models. Predictive models were generated with SW-kNN MFA. The most significant model 1 is having internal predictivity 64.24% (q2 = 64.24) and external predictivity 61.57 % (pred_r2 = 0.61.57). Model showed that steric (S_584), and electrostatic (E-295) interactions play important role in determining DPP IV inhibitory activity.
Select your language of interest to view the total content in your interested language
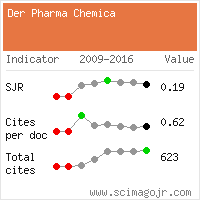
Google Scholar citation report
Citations : 15261
Der Pharma Chemica received 15261 citations as per Google Scholar report
Der Pharma Chemica peer review process verified at publons