Research Article - Der Pharma Chemica ( 2024) Volume 16, Issue 1
Exploring the Potential Flavonoids as an Anti-Rheumatoid Arthritis Agent: An In silico Studies
Abhilasha Singh1, Sharmistha Banerjee2, Surada Prakash Rao3, Ashish Majumdar3 and Madhuri Baghel4*2Department of Pharmaceutical Sciences, Chhattisgarh Swami Vivekanand Technical University, Newai Bhilai, India
3Department of Pharmaceutical Sciences, Columbia Institute of Pharmacy, Raipur, India
4Department of Pharmaceutical Sciences, Apollo College of Pharmacy, Anjora, India
Madhuri Baghel, Department of Pharmaceutical Sciences, Apollo College of Pharmacy, Anjora, India, Email: banchhormadhuri@gmail.com
Received: 22-Dec-2023, Manuscript No. DPC-23-123311; Editor assigned: 27-Dec-2023, Pre QC No. DPC-23-123311 (PQ); Reviewed: 10-Jan-2024, QC No. DPC-23-123311; Revised: 29-Jan-2024, Manuscript No. DPC-23-123311 (R); Published: 26-Feb-2024, DOI: 10.4172/0975-413X.16.1.204-210
Abstract
Rheumatoid arthritis is an autoimmune disease which causes systemic complications, disability and even death. Its pathophysiology involves chronic inflammation of synovial membrane. New molecules in the biologic pathway have been discovered and are considered new target for experiments. In this experiment 100 flavonoids showing anti-inflammatory activities in different studies were evaluated using in silico approach to get the best possible lead. Orientin (IFD score: 640.28) and leucocyanidin (IFD score: 638.22) showed highest binding affinity with protein 4K6Z, while vitexin (IFD score: 622.21) and hyperoside (IFD score: 623.55) showed highest binding affinity with protein 4G1W among the set of flavonoids taken. To support the results of this investigation, leucocyanidin and orientin could be investigated further for the treatment of rheumatoid arthritis through both in vitro and in vivo experiments.
Keywords
Rheumatoid arthritis; Induced fit docking; Anti-inflammatory; Flavonoids; In silico
Introduction
Rheumatoid Arthritis (RA) is an autoimmune inflammatory disease which is characterized by synovitis (synovium of a joint becomes swollen or inflamed). It is accompanied by multi-organ disorders, in addition to pain, multiple joints are affected and stiffness occurs. Destruction of joint progresses rapidly after onset, because of physical deformation and dysfunction which is irreversible in nature [1]. Currently, it has been reported that around 40% of RA patients who are not treated well cannot work within 10 years of diagnosis [2]. Other pathological symptoms may appear that includes pleuritic, pericarditis, small vessel vasculitis, pulmonary granulomas and keratitis [3]. Various approaches have been adopted to treat or reduce the ill effects of RA. The most commonly used method is anti-inflammatory related treatment. Reportedly, Non-Steroidal Anti- Inflammatory Drugs (NSAIDS) and corticosteroids are most promising agents for efficiently reducing the pain and stiffness in joints, without subsiding the progression of disease [4]. Traditional treatment procedures cause undesirable side effects, while some of them can even worsen the disease status. In this regard, secondary metabolites or bio actives secreted from plants particularly alkaloids, saponins and flavonoids can slow down the progression of disease or because of its anti-inflammatory, antioxidant, enzymatic and immunomodulatory activities. Thus, these secondary metabolites are becoming suitable candidates for therapeutic development which in turn will improve the life of RA [5]. Flavonoids are becoming an interesting candidate to be used as anti-inflammatory agents against RA. Flavonoids are known for inhibiting the production of Nitric Oxide (NO), pro-inflammatory cytokines and eicosanoids that interfere with the NF-kB transcription factor [6-7]. Using molecular docking approach, flavonoids were evaluated for their binding interactions with two protein receptors (namely MAPK8-Pdb id 4G1W and JAK1-Pdb id 4K6Z). Initially, SP (Standard Precision) docking was used to shortlist the top 10 flavonoids, further after XP (extra precision) docking, top two ligands that exhibited the highest binding affinity and interaction with the target protein, surpassing even the reference compound Upadactinib, were subjected to IFD (Induced Fit Docking), a powerful and accurate docking method. IFD generated various binding poses and assign corresponding binding scores.
Materials and Methods
Protein preparation
Two protein targets were selected based on literature reviews showing signal transduction network in the inflammatory process of RA (Table 1). The three-dimensional crystallographic structure of human tyrosine-protein kinase JAK1 (PDB ID: 4K6Z) and MAPK8 (PDB ID:4G1W) were downloaded from protein data bank. Both the proteins were then prepared and processed by protein preparation wizard (in Maestro Schrodinger Suite) [8-10] (v13.1) after applying custom preset, in pre-process method all water molecules were deleted in range of 3.00 Å. Extra chains were removed and structures were refined and minimized using the Optimized Potentials for Liquid Simulations force field (OPLS_2005).
| S. No. | Protein Target | PDB ID | Comment | Structure |
|---|---|---|---|---|
| 1 | Tyrosine-protein kinase JAK1 | 4K6Z | Organism(s): Homo sapiens mutation: No ID resolution: 2.73 Å | 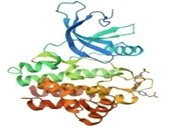 |
| 2 | MAPK8 | 4G1W | Organism: Homo sapiens mutation: No resolution: 2.45 Å | 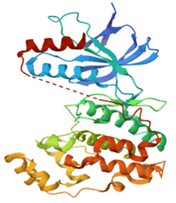 |
Table 1: Protein target taken for docking in rheumatoid arthritis.
Selection of compound and ligand preparation
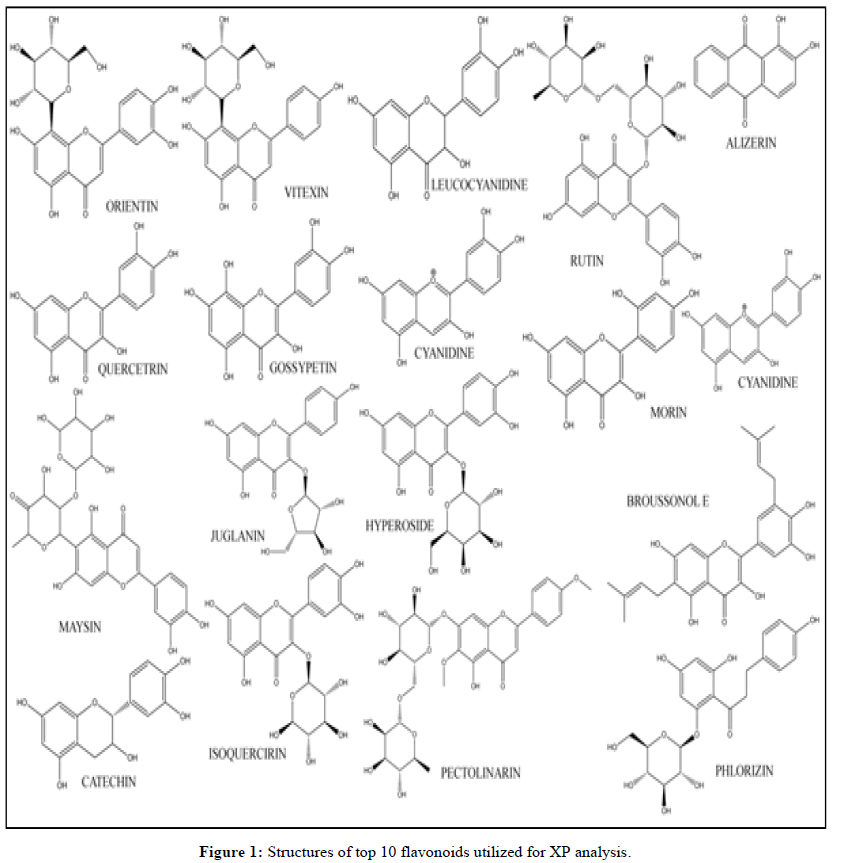
With help of literature survey 100 flavonoids were identified showing anti-inflammatory and anti-oxidative properties from literature review. The selected compounds were downloaded from the PubChem database, in SDF format. With the help Maestro Schrodinger (13.1) LigPrep wizard the minimized 3D structures were prepared within pH range of 7.0 ± 2.0. Further minimization using the OPLS_2005 force field generated best possible stereoisomers of ligands depending on chiral centers present in each molecule. The structures of flavonoids (top 10 for each protein target) utilized for XP docking are illustrated in Figure 1.
Receptor grid generation
Grid generation create a grid file which shows the active site of receptor for glide ligand docking. It helps the ligand to bind specifically within a particular area of receptor protein during docking. Glide determines a default center and a default size for the region for which grids will be calculated using the position and size of the ligand. Both receptor proteins grid was generated and processed by OPLS_2005 force field for the minimized structure in glide.
Glide SP (standard precision) and XP (extra precision) docking
SP glide docking of 100 ligands prepared under LigPrep was performed with grid generated receptor protein (4K6Z and 4G1W). The charge cut-off and Vander Waals radius scaling factor was set to 0.15 and 0.80 respectively. The docking score generated by SP glide docking helps us in analyzing ligand receptor interaction and best possible poses. Top ten flavonoids were selected based on SP glide docking score and subjected to XP ligand docking which usually is more accurate in predicting the binding affinity of ligands with the receptor (Figure 1).
Induced Fit Docking (IFD)
Two compounds showing maximum interaction as of reference compound and with top docking score were selected and subjected to IFD, which is a more rigorous scoring method and generate more accurate poses of the ligands with protein receptor. OPLS_2005 force field was applied after generating grid around the co-crystallized ligand of the receptor [11]. Residues within two Å were refined and top five best possible poses of ligands were selected based on the IFD score.
Results and Discussion
Mitogen activated protein kinases or MAPKs, have been shown to be important regulators of the generation of pro-inflammatory cytokines and subsequent signaling events that cause inflammation and damage to joints. Hence MAPK (4G1W) is an attractive therapeutic target and has a potential therapeutic avenue for RA [12]. Similarly Janus kinase (JAK, 4K6Z) inhibitors are potential target for treatment of RA. These are the newest category of disease modifying drugs to be developed for the treatment of RA [13]. Through rigorous literature survey it was found that various flavonoids show anti- inflammatory and anti-oxidant activities which can be helpful in treatment and management of rheumatoid arthritis. Hundred flavonoids molecule showing anti-inflammatory action were downloaded from PubChem and saved in SDF format. These hundred compounds were further subjected to SP and XP Docking and from top ten compounds based on docking score two compounds having maximum interaction with protein target as of reference compound were selected for IFD. Results of top ten ligands obtained after analyzing XP glide docking score are enlisted in Table 2 (protein target- 4K6Z) and Table 3 (protein target- 4G1W). Tables 4 and 5 show IFD score and the interaction of ligand with protein receptor 4K6Z and 4G1W respectively [14-26].
| S. No | Pubchem ID | Flavonoids | Docking score | Mol weight (g/mol) | |
|---|---|---|---|---|---|
| SP | XP | ||||
| 1 | 5281675 | Orientin | -9.6238 | -14.277 | 448.4 |
| 2 | 5280441 | Vitexin | -8.8513 | -12.333 | 432.4 |
| 3 | 71629 | Leucocyanidin | -8.8393 | -11.289 | 306.27 |
| 4 | 5280805 | Rutin | -9.8654 | -11.048 | 610.5 |
| 5 | 5280459 | Quercitrin | -8.8201 | -10.636 | 448.4 |
| 6 | 5280647 | Gossypetin | -8.4622 | -10.331 | 318.23 |
| 7 | 128861 | Cyanidin | -9.0574 | -10.287 | 287.24 |
| 8 | 194340 | Maysin | -8.6242 | -10.04 | 576.5 |
| 9 | 5281670 | Morin | -8.8582 | -9.944 | 302.23 |
| 10 | 5318717 | Juglanin | -8.4556 | -9.6972 | 418.3 |
| 11 | 5.9E+07 | Upadactinib | -8.1702 | -7.094 | 380.4 |
Table 2: SP and XP docking score of top ten ligands (protein target: 4K6Z).
| S. No | Pubchem ID | Flavonoids | Docking score | Mol weight (g/mol) | |
|---|---|---|---|---|---|
| SP | XP | ||||
| 1 | 5280441 | Vitexin | -8.9859 | -13.626 | 432.4 |
| 2 | 5280805 | Rutin | -8.7085 | -13.531 | 610.5 |
| 3 | 168849 | Pectolinarin | -8.7035 | -12.593 | 622.6 |
| 4 | 5281643 | Hyperoside | -8.8636 | -12.006 | 464.4 |
| 5 | 5280804 | Isoquercitrin | -8.6078 | -11.537 | 464.4 |
| 6 | 1E+07 | Broussonol E | -8.1161 | -10.963 | 438.5 |
| 7 | 9064 | Catechin | -8.1355 | -9.8645 | 290.27 |
| 8 | 6072 | Phlorizin | -8.3587 | -9.5285 | 436.4 |
| 9 | 6293 | Alizarin | -8.1253 | -9.4655 | 240.21 |
| 10 | 128861 | Cyanidin | -8.3592 | -9.2688 | 287.24 |
| 11 | 126941 | Upadactinib (Reference) | -8.1139 | -8.074 | 454.4 |
Table 3: SP and XP docking score of top ten ligands (protein target: 4G1W).
| Pubchem ID | IFD score (Top 5 poses) | XP score | Interacting residues | Type of interaction |
|---|---|---|---|---|
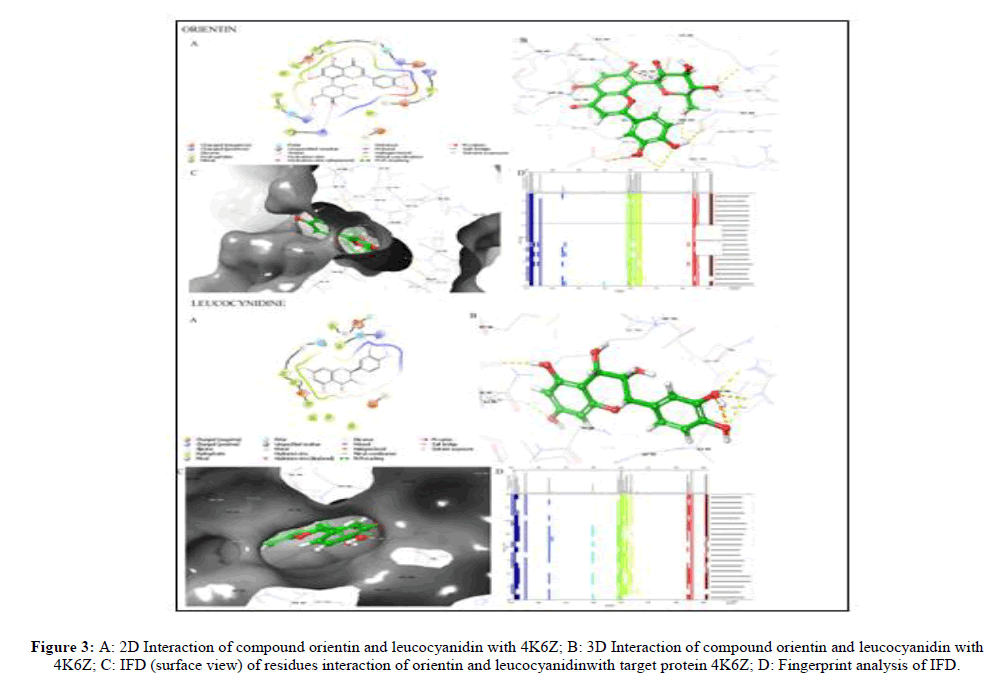
| 5281675 | 640.28 | -14.2771 | ARG 1007 | Hydrogen bond |
| (Orientin) | 638.43 | LEU 959 | Hydrogen bond | |
| 637.43 | GLU 957 | Hydrogen bond | ||
| 636.78 | GLY 1020 | Hydrogen bond | ||
| 636.71 | GLU 883 | Hydrogen bond | ||
| PRO 960 | Hydrogen bond | |||
| ARG 879 | Hydrogen bond | |||
| 71629 (Leucocyanidin) | 638.22 | -11.2892 | ARG 1007 | Hydrogen bond |
| 638.14 | LEU 959 | Hydrogen bond | ||
| 637.94 | GLU 957 | Hydrogen bond | ||
| 637.67 | GLU 883 | Hydrogen bond | ||
| 637.46 | GLY 1020 | Hydrogen bond |
Table 4: Result of best performed ligand molecules with IFD score (top five poses), type of interactions, interacting amino acid residues for Protein target 4K6Z.
| Pubchem ID | IFD score (Top 5 poses) | XP score | Interacting residues | Type of Interaction |
|---|---|---|---|---|
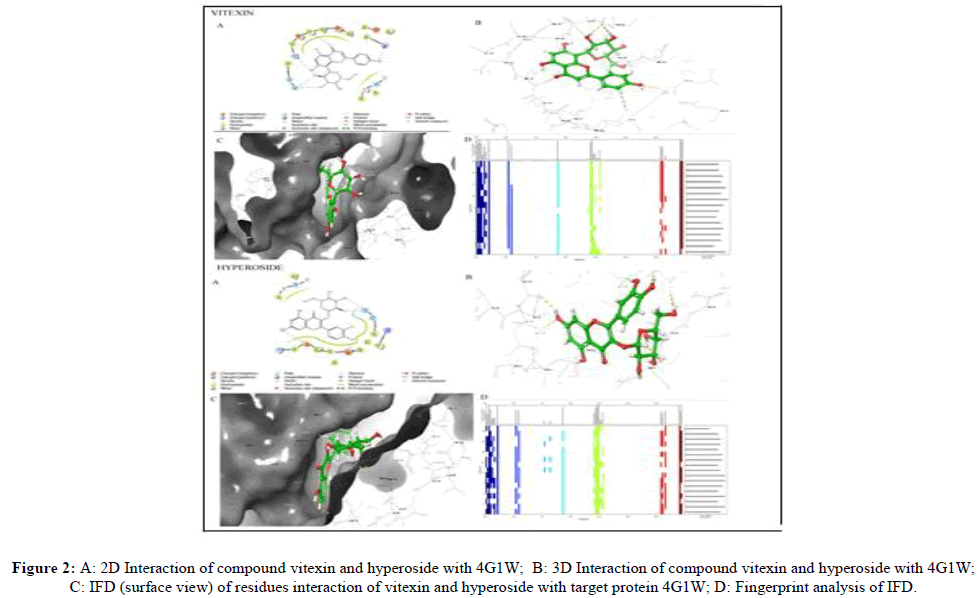
| 5280441 (Vitexin) | 622.21 | -13.626 | SER 155 ASN 114 MET 111 LYS 55 |
Hydrogen bond Hydrogen bond Hydrogen bond Hydrogen bond |
| 619.98 | ||||
| 619.88 | ||||
| 619.66 | ||||
| 619.65 | ||||
| 5281643 (Hyperoside) | 623.55 | -12.006 | ASP 112 GLU 109 SER 155 |
Hydrogen bond Hydrogen bond Hydrogen bond |
| 622.64 | ||||
| 622.28 | ||||
| 622.28 | ||||
| 621.95 |
Table 5: Result of best performed ligand molecules with IFD score (top five poses), type of interactions, interaction amino acid residues for protein target 4G1W.
Orientin and leucocyanidine interacted efficiently with protein 4G1W with highest binding energy of -14.2771 kcalmol-1 and -12.3327 kcalmol-1 respectively, which is higher than reference upadacitinib (-8.074 kcalmol-1). Similarly for protein 4K6Z vitexin and hyperoside had higher binding score (-13.6202 kcalmol-1 and -12.0056 kcalmol-1) than the Upadacitinib (-7.094 kcalmol-1). All top 10 flavonoids have higher binding score than the reference. Orientin (IFD score: 640.28) and leucocyanidin (IFD score: 638.22) showed highest binding affinity with protein 4K6Z, while vitexin (IFD score: 622.21) and Hyperoside (IFD score: 623.55) showed highest binding affinity with Protein 4G1W among the set of flavonoids taken.
With protein 4G1W, Upadacitinib formed two hydrogen bonds with amino acid residue MET111 (with carboxyl) and ASN114 (with amino group). Vitexin showed four hydrogen bonds, interaction with two amino acid residues were similar (MET111 and ASN114) and additional interactions were with SER155 and LYS55. Hyperoside formed three hydrogen bonds with ASP112, GLU109 and SER155. 2D, 3D interaction, IFD and Fingerprint analysis of IFD for protein 4G1W are illustrated in Figure 2.
With protein 4K6Z upadacitinib formed three hydrogen bonds with the amino acid residues ARG1007 and LEU959, with LEU 959 it formed two bridged H-bonds. Orientin formed seven hydrogen bonds, out of which two interactions were similar with the reference (with ARG100, LEU959), other interactions were with amino acid residues GLU957, GLY1010, GLU883, PRO960 and ARG879. Leucocyanidine formed five hydrogen bonds, out of which two were similar (with ARG100, LEU959). Other interactions were with GLU957, GLY1010 and GLU883 that were similar with the orientin. 2D, 3D interaction, IFD and fingerprint analysis of IFD for protein 4K6Z are illustrated in Figure 3.
Conclusion
The results showed that among the set of 100 flavonoids, orientin, leucocyanidine, vitexin and hyperoside displayed excellent antirheumatoid arthritis effects in silico. The results of molecular docking also suggest that orientin, leucocyanidine form strong protein ligand complexes with the protein target 4K6Z while vitexin and hyperoside form strong protein ligand complexes with the protein target 4G1W. Current research indicates that these phyto compounds have the potential to be further developed through chemical synthesis and biological activity investigation. Although phyto compounds may require additional enhancement of their binding affinity prior to their potential application as therapeutic candidates, this thorough analysis may encourage more research into the development of possible derivatives derived from these phyto compounds to the targets found in arthritis, which may eventually serve to replace the currently available drugs utilized for treatment of RA.
Declaration of Interest Statement
The authors have no conflicts of interest to declare.
References
- Tanaka Y. Inflamm Regen. 2023; 43(1): p. 3-20.
[Crossref] [Google Scholar] [PubMed]
- Deighton C, Mahony R, Tosh J, et al. Bmj. 2009; 16: p. 338.
[Crossref] [Google Scholar] [PubMed]
- Guo Q, Wang Y, Xu D, et al. Bone Res. 2018; 6(1): p. 15.
[Crossref] [Google Scholar] [PubMed]
- Mrid RB, Bouchmaa N, Ainani H, et al. Biomed Pharmacother. 2022; 151(86): p. 113-126.
[Crossref] [Google Scholar] [PubMed]
- Santiago LA, Neto RN, Santos Ataide AC, et al. Clin Phytoscience. 2021; 7(1): p. 1-10.
- Burda S, Oleszek W. J Agric Food Chem. 2001; 49(6): p. 2774-2779.
[Crossref] [Google Scholar] [PubMed]
- Li AN, Li S, Zhang YJ, et al. Nutrients. 2014; 6(12): p. 6020-6047.
[Crossref] [Google Scholar] [PubMed]
- Gou KJ, Zeng R, Ren XD, et al. Immunol Lett. 2018; 201: p. 59-69.
[Crossref] [Google Scholar] [PubMed]
- Song X, Zhang Y, Dai E, et al. Int Immunopharmacol. 2020; 80: p. 106-179.
[Crossref] [Google Scholar] [PubMed]
- Thalhamer T, McGrath MA, Harnett MM. Rheumatology. 2008; 47(4):p. 409-114.
[Crossref] [Google Scholar] [PubMed]
- Harrington R, Al Nokhatha SA, Conway R. J Inflamm Res. 2020; 23(5): p. 519-531.
[Crossref] [Google Scholar] [PubMed]
- Subbiah V, Xie C, Dunshea FR, et al. Food Rev Int. 2023; 39(8): p. 5786-5813.
- Augustine C. J Struct Chem. 2019; 60: p. 198-209.
- Guardia T, Rotelli AE, Juarez AO, et al. Il farmaco. 2001; 56(9): p. 683-687.
[Crossref] [Google Scholar] [PubMed]
- Ginting CN, Lister IN, Girsang E, et al. J Phys Conf Ser. 2019; 1374(1): p. 12-33.
- Emam M, El Raey MA, El-Haddad AE, et al M. Molecules. 2018; 24(1): p. 138.
[Crossref] [Google Scholar] [PubMed]
- Yoshida T, Wang H, Nair MG, et al. J Nat Prod. 1999; 62: p. 802.
- Song Y, Kim HD, Lee MK, et al. PloS one. 2017; 12(1): p. 169-509.
[Crossref] [Google Scholar] [PubMed]
- Dos Santos Lima B, de Alcantara Campos C, Da Silva Santos AC, et al. Food Chem Toxicol. 2019; 126:15-24.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Zhang C, Wang X, Huo S. Artif Cells Nanomed Biotechnol. 2019; 47(1): p. 3614-3620.
[Crossref] [Google Scholar] [PubMed]
- Li X, Wang Y. Inflammation. 2020; 43(5): p. 1729-1741.
[Crossref] [Google Scholar] [PubMed]
- Sun K, Luo J, Guo J, et al. Osteoarthr Cartil. 2020; 28(4): p. 400-409.
[Crossref] [Google Scholar] [PubMed]
- Kim EH, Lee S, Chung MJ. Korean J Food Preserv. 2021; 28(7): p. 1000-1009.
- Ryu HW, Park MH, Kwon OK, et al. Bioorg Chem. 2019; 92: p. 103-233.
[Crossref] [Google Scholar] [PubMed]
- Li T, Li F, Liu X, et al. Phytother Res. 2019; 33(3): p. 756-767.
[Crossref] [Google Scholar] [PubMed]
- Jin Y, Zhou J, Zhao X, et al. J Mater Chem. 2022; 18(7): p. 2759-2774.
[Crossref] [Google Scholar] [PubMed]