Research Article - Der Pharma Chemica ( 2018) Volume 10, Issue 10
New Aliphatic Alcohols from Seeds of Momordica charantia Linn. and Prediction of their Biological Activity
Deepti Katiyar1*, Vijender Singh2, Mohammed Ali3
1Department of Pharmacognosy, KIET School of Pharmacy, Ghaziabad, UP, India
2School of Pharmacy, Sharda University, Greater Noida, UP, India
3Department of Pharmacognosy and Phytochemistry, Faculty of Pharmacy, Jamia Hamdard, New Delhi, India
- *Corresponding Author:
- Deepti Katiyar
Department of Pharmacognosy
KIET School of Pharmacy
Ghaziabad, UP, India
Abstract
Momordica charantia is a very well-known traditional medicinal herb. The current research investigation aimed towards the isolation, characterization and prediction of biological activity spectra of the phytoconstituents from the seeds of Momordica charantia Linn. (Cucurbitaceae). The Ethanolic extract of Momordica charantia seeds led to isolate two new aliphatic alcohols: n-nonatriacontan-19α-ol &n-henetetracontan-19α-ol and four known compounds: n-tetracosane, n-pentacosane, n-pentatriacontane, n-heptatriacontan-15α-ol. These isolated phytomolecules can act as the marker compounds in order to establish the identity, quality and purity of the drug. The observations of the in silico profiling of these compounds shall be very useful for the upcoming reaearchers for establishing them as pharmacologically active moieties.
Keywords
n-nonatriacontan-19α-ol, n-henetetracontan-19α-ol, n-tetracosane, n-pentacosane, n-pentatriacontane, n-heptatriacontan-15α-ol.
Introduction
Momordica charantia Linn. (Family: Cucurbitaceae) is commonly known as karela in hindi; bitter gourd, bitter cucumber, bitter melon or balsam pear in English and karavella in Sanskrit [1]. It is a monoecious annual climber which grows upto 5 m in height cultivated in East Africa, South America, China, Malaya and India upto an altitude of 1500 m and it also grows wildely in tropical and sub-tropical Africa, Asia, America and Caribbean [2]. The plant possesses thin, branched, grooved and twisted stem; glabrous leaves with margin containing sharp protruding points; solitary, unisexual, pale yellow to orange flowers; dark green oblong, 5-25 cm long pepo fruits with compressed brownish 12-16 mm long seeds embedded in red pulp [3].
The Ayurvedic Pharmacopoeia mentions the medicinal uses of Momordicacharantia as Laghu (easily digestible), Bhedana (purgative), Javara (fever), Tikta (bitter), Asradosa (dyscarasia), Prameha (obstinate urinary diseases including diabetes), Paandu (anaemia) and Krimi (worm infestation) [4]. Various recent pharmacological studies have displayed the medicinal potential of M. charantia as anti-oxidant, anti-viral, anti-bacterial, anti-HIV, anti-cancer, anti-diabetic and anthelmintic. It is also used in hypertension, obesity and modulating immune system [2,5-8]. More than 220 phytoconstituents have been isolated from leaves, fruits, stems, entire plant, pericarp, callus tissues and seeds [9] which include glycosides, alkaloids, terpenoids, fixed oils, steroids, proteins, inorganic compounds, monoterpenes, carbohydrates, benzanoids, sesquiterpenes etc. [8].
Cucurbitacins, momordicins, cucurbitanes, momordenol, cucurbitins, momordicilin, cycloartenols, galactouronic acids, goyaglycosides, multiflorenol, diosgenin, gallic acid etc. have been identified as the major constituents of Momordica charantia [9-11]. The major active hypoglycemic constituent is charantin also known as momorcharin which is a 1: 1 mixture of β-sitosterol β-D-glucoside and stigmastandienol β-D-glucoside [12]. Anti-diabetic effects have also been shown due to some other constituents like v-insulin, polypeptide-p, p-insulin (insulin like action) [13-15]; momordin (activates PPAR β/δ) [16]; vicine (hypoglycemic effect) [17] and momordicoside S and T (enhances glucose clearance and basal metabolic rate) [18].
Materials and Methods
General chemical procedures
All melting points (mp) were determined in centigrade scale in one–end open capillary on a thermoelectrical melting point apparatus. The IR spectra were measured on IR affinity-1 Fourier transform infrared spectrometer model (Schimadzu). The mass spectra were recorded on a JEOL-Accu TOF (time of flight) JMS-T100LC mass spectrometer having a DART (direct analysis in real time) source. The m/z (mass to charge ratio) values of the more intense peaks are mentioned and the figure in a bracket attached to each m/z values indicated relative intensities with respect to the base peak. The 1H and 13C-NMR spectra were scanned on BrukerAvIII HD-300 and 75 MHz, respectively, an instrument in CDCl3 and MeOD solvents using TMS as an internal standard. The coupling constants (J values) are expressed in Hertz (Hz). Column chromatography was performed on a silica gel (60-120 mesh; Qualigen, Mumbai, India) column. TLC (Thin Layer Chromatography) was run on silica gel G 60 F 254 (Qualigen) coated aluminium sheets. Spots were visualised by exposing to iodine vapors, UV (ultraviolet) radiation and spraying with ceric sulfate solution.
Plant material
The dried seeds of Momordica charantia were procured from KhariBaoli market, Delhi. They were authenticated by Dr. SunitaGarg, Chief Scientist, Raw Material Herbarium and Museum, Delhi (RHMD), CSIR-NISCAIR, New Delhi and assigned the voucher specimen/Accession number (Ref No. NISCAIR/RHMD/Consult/ 2015/ 2911/ 104-1). The voucher specimen is preserved in the herbarium section of Department of Pharmacognosy, KIET School of Pharmacy, Ghaziabad, Uttar Pradesh.
Extraction
The dried seeds of Momordica charantia (4.0 kg) were crushed to smaller pieces, re-dried, coarsely powdered, defatted with petroleum ether and then extracted in ethanol (95%) using Soxhlet Apparatus for 72 h. The extract was then concentrated under reduced pressureto yield 270.93 g (6.77%) of dark reddish brown orange mass.
Preparation of slurry
The concentrated alcoholic extract of Momordica charantia (45 g) was taken in a china dish, placed on water bath and methanol was added in fractions to attain the desired consistency. Silica gel for column chromatography (60-120 mesh) was added gradually with constant mixing with a stainless steel spatula to obtain the slurry required to be packed into the column. Then, it was dried in air and the larger lumps were broken up. Finally, the uniform particle size was obtained by passing it through a sieve (No. 8).
Isolation of phytoconstituents
The adsorbent cotton was plugged in the lower end of a dry column (5 feet high, 2.5 inches diameter) and then it was filled with petroleum ether upto half of its height. The required length of the packed column was obtained by adding silica gel in small proportions and allowing it to settle down. The blank column was run thrice with solvent to allow the bubbles to escape. Then the column was packed with dried silica gel slurry of the extract and the column was plugged with adsorbent cotton. Different solvents (2.5 l each) with increasing polarity were used to elute the column. The column was developed and eluted with solvents of increasing polarity in various combinations, viz., petroleumether, chloroform in petroleum ether (0.5%, 1%, 2%, 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%) chloroform (100%), and methanol in chloroform. The Thin layer Chromatography was performed for the fractions (250 ml each) thus collected. The fractions exhibiting the similar chromatographic profile were combined and concentrated.
PASS prediction
In this molecular modelling study, structures were generated with the aid of Chem3D Ultra-9.00 and HyperChem-v.6.02, Wolfram Research Mathematics 6.0 software. Lone pairs of electrons and hydrogen atoms were added where appropriated. The equilibrium geometries of compounds were located using MM+(for Hyper Chem) and MM2 (for Chem3D) functional set. In the next step, RHF calculation (semiempirical AM1 method, the self-consistent field of Hartree-Fock) were performed and bond length, angles, torsion angles and partial charges have been calculated. Calculations were performed on a Intel (R) Core2 (TM) CPU 6600 @ 2.4 GHz Pentium IV computer with 2 GB RAM.
Results and Discussions
Isolation and characterization of phytoconstituents
Compound MC-1
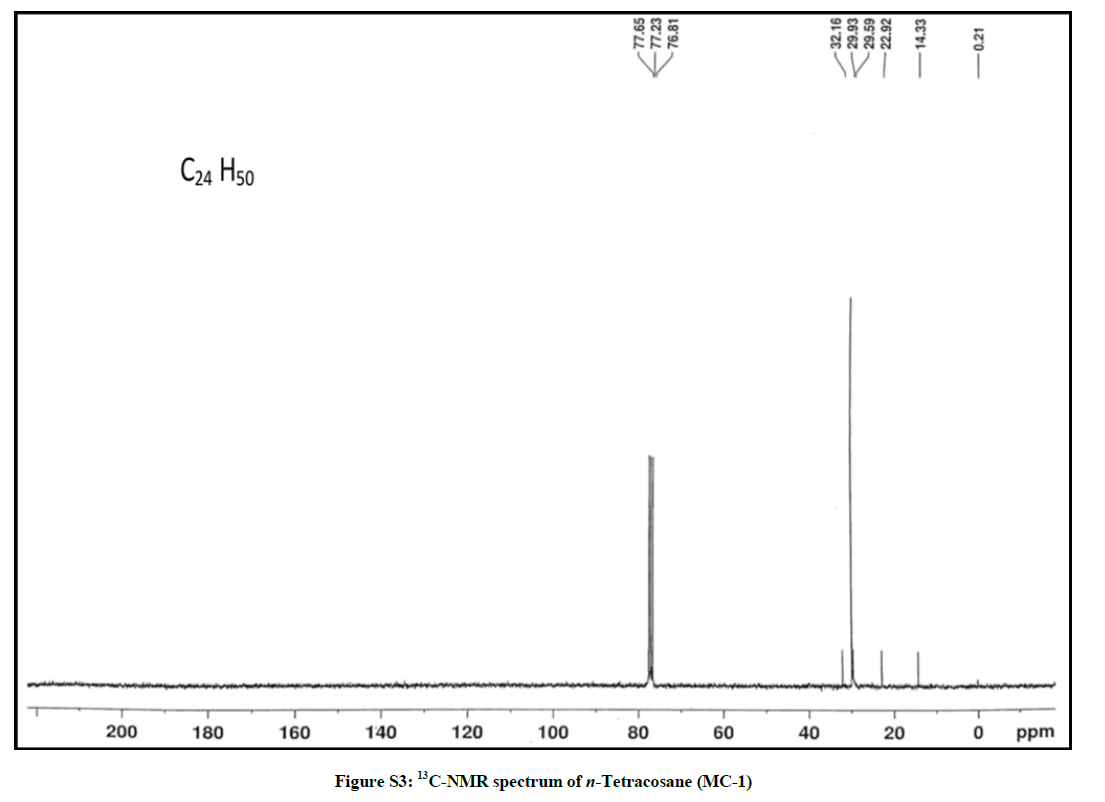
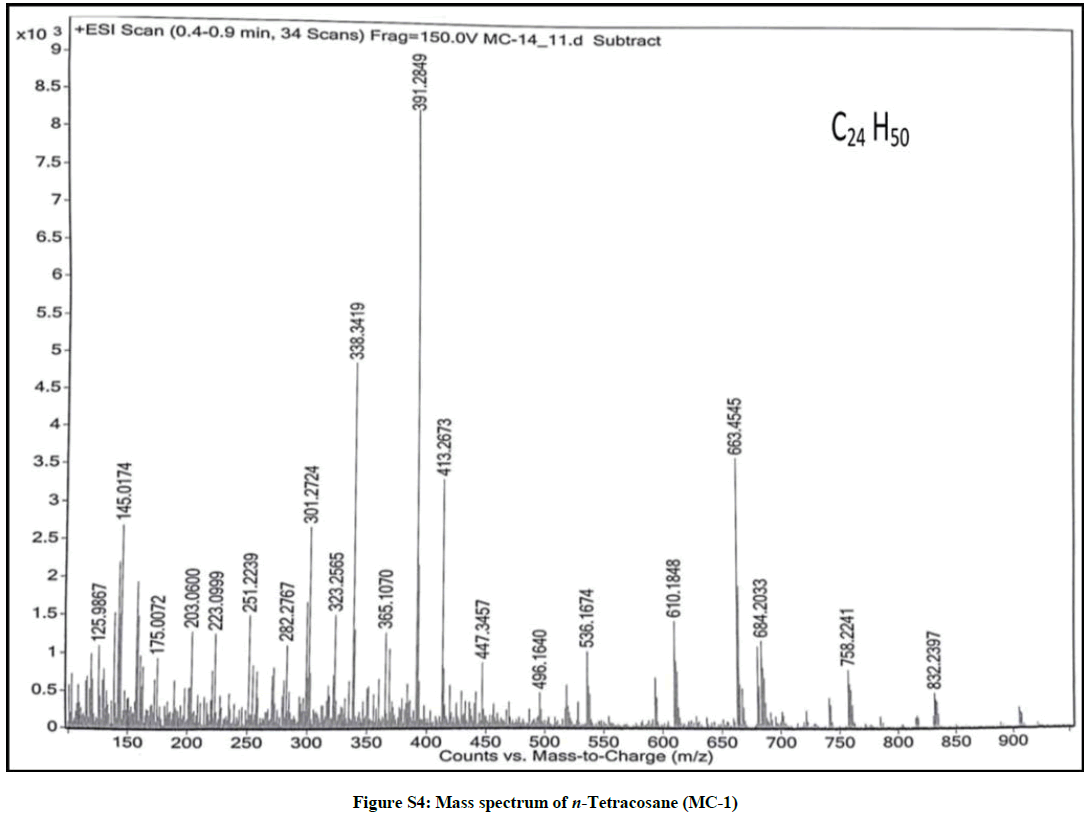
Elution of the column with petroleum ether: chloroform (90: 10) yielded colorless powder of MC-1, 85 mg (0.04% yield). Rf: 0.80 (petroleum ether: chloroform: methanol, 2: 4: 0.1). Melting point: 71ºC-72ºC; IR υmax (KBr): 2926, 2854, 1466, 1215, 1077, 929, 757 cm-1; 1H-NMR (CDCl3): δ (ppm)=1.56 (2H, m, CH2), 1.33 (2H, m, CH2), 1.31 (2H, m, CH2), 1.28 (4H, m, 2α CH2), 1.25 (34 H, brs, 17 × CH2), 0.88 (3H, t, J=6.0 Hz, Me-1), 0.85 (3H, t, J=7.5 Hz, Me-24); 13C-NMR (CDCl3): δ (ppm)=32.16 (CH2), 29.93 (19 × CH2), 29.59 (CH2), 22.92 (CH2), 14.33 (Me-1, Me-24). ESI MS m/z (rel. int.): 338 [M]+(C24H50) (50.3).
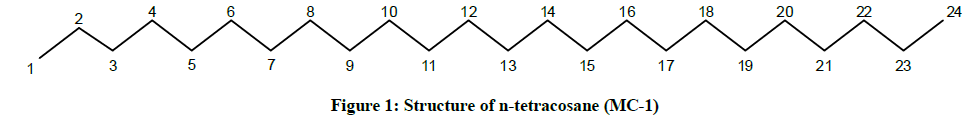
The IR spectrum of compound MC-1 exhibited an absorption band for long chain hydrocarbon at 757 cm-1. The absence of any distinctive band in the range 3500-3200 cm-1 and 1750-1600 cm-1 suggested the saturated nature of the molecule without hydroxyl, amino, carboxylic, carbonyl and other functional groups. The mass spectrum had a molecular ion peak at m/z 338 consistent with the molecular formula of a saturated hydrocarbon, C24H50. The absence of [M – Me]+ ion peak indicated the straight chain nature of the molecule. The 1H-NMR spectrum of MC-1 showed two three - proton triplets at δ 0.88 (J=6.0 Hz) and 0.85 (J=7.5 Hz) ascribed to the terminal C-1 and C-24 primary methyl protons, respectively. Three two-proton multiplets at δ 1.56, 1.33 and 1.31, a four-proton multiplet at δ 1.28 and a broad singlet at δ 1.25 (34 H) were associated with the methylene protons. The 13C-NMR spectrum of MC-1 displayed signals at δ 14.33 due to presence of methyl carbons C-1 and C-24, and between δ 32.16-22.50 assigned to the methylene carbons. The absence of any signal beyond δ 1.56 in the 1H-NMR spectrum and δ 32.16 in the 13C-NMR spectrum ruled out the unsaturated nature of the molecule and existence of any functional group in it. On the basis of foregoing spectral data analysis, the structure of MC-1 has been elucidated as n-tetracosane (Figure 1).
Compound MC-2
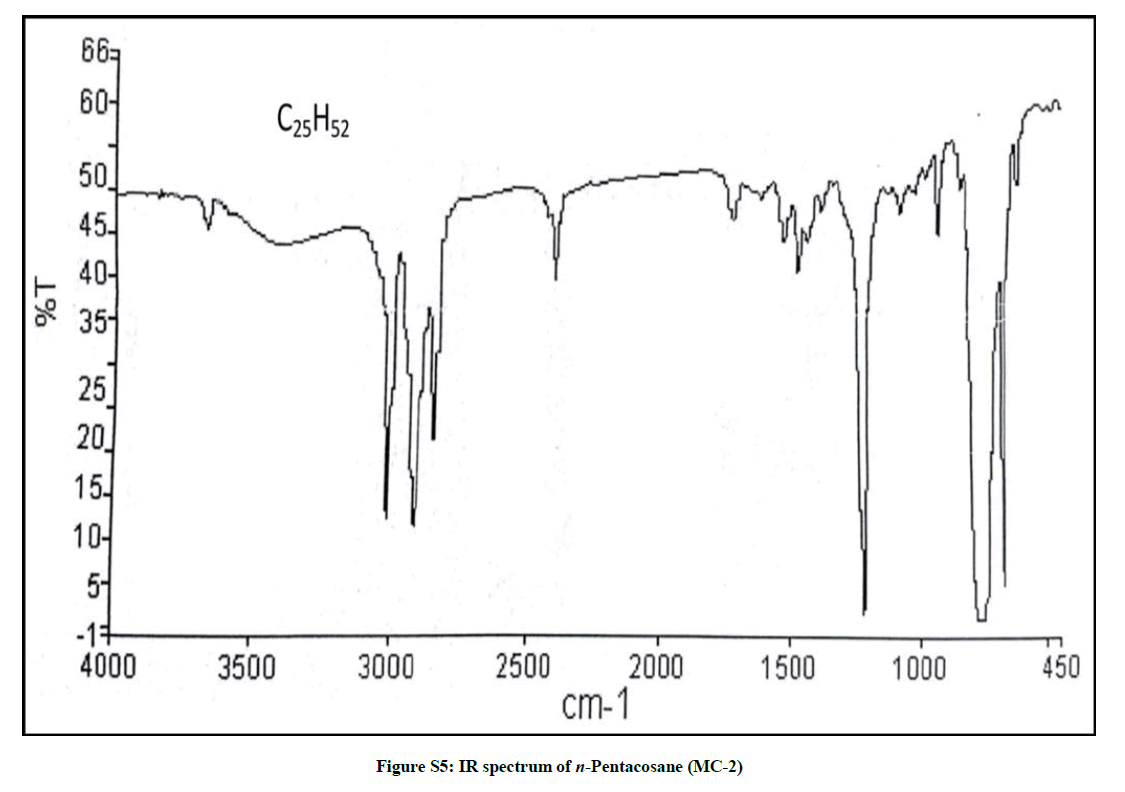
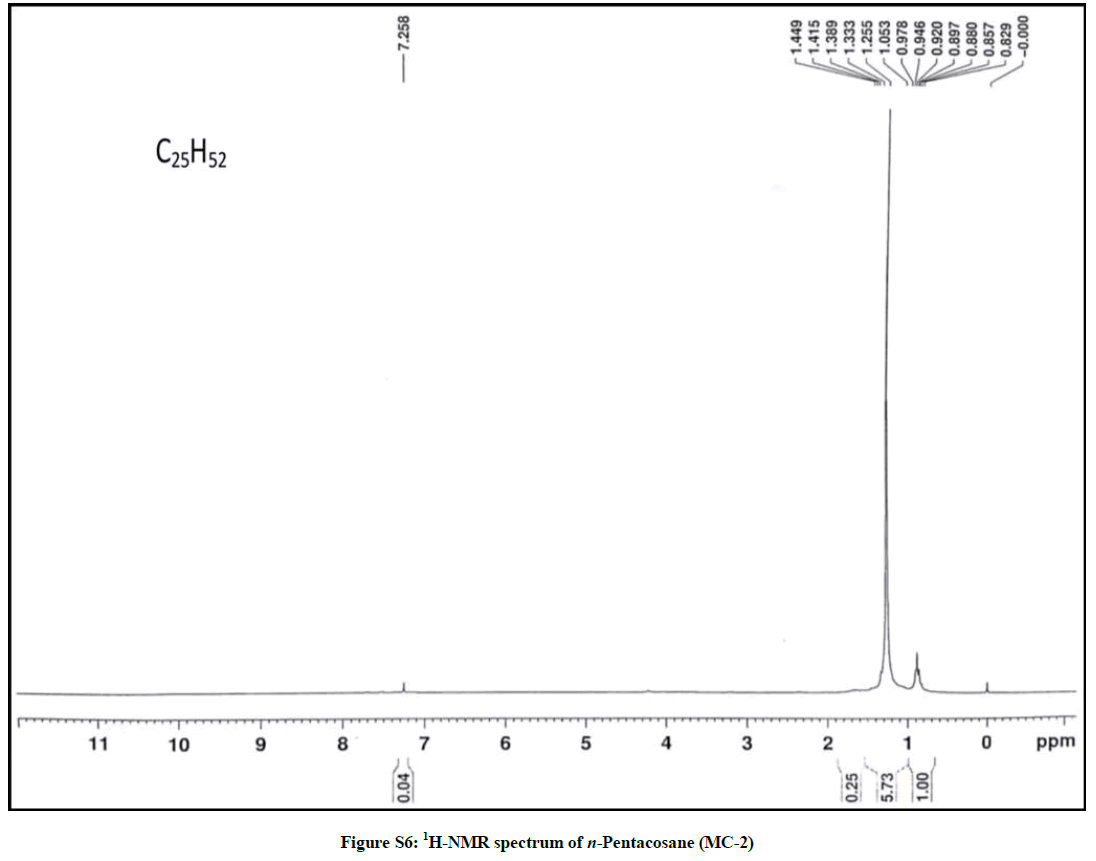
Elution of the column with petroleum ether: chloroform (80: 20) afforded colorless powder of MC-2, 97 mg (0.045% yield). Rf: 0.82 (benzene: chloroform: methanol, 2.5: 4.5: 0.6). Melting point: 78ºC-79ºC. IR υmax (KBr): 2927, 2854, 1466, 1378, 1215, 1078, 849, 757 cm-1; 1H-NMR (CDCl3): δ (ppm)=1.64 (2H, m, CH2), 1.50 (2H, m, CH2), 1.45 (2H, m, CH2), 1.41 (2H, m, CH2), 1.38 (2H, m, CH2), 1.33 (2H, m, CH2), 1.29 (6H, s, 3 × CH2), 1.25 (28H, brs, 14 × CH2), 0.89 (3H, t, J=6.0 Hz, Me-1), 0.85 (3H, t, J=7.2 Hz, Me-25); 13C-NMR (CDCl3): δ (ppm)=32.16 (CH2), 29.93 (20 × CH2), 29.59 (CH2), 22.92 (CH2), 14.33 (Me-1, Me-25). ESI MS m/z (rel. int.): 352 [M]+(C25H52O) (2.1).
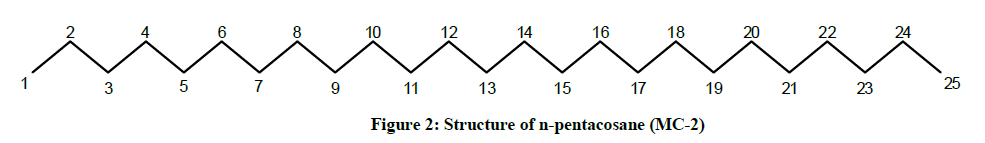
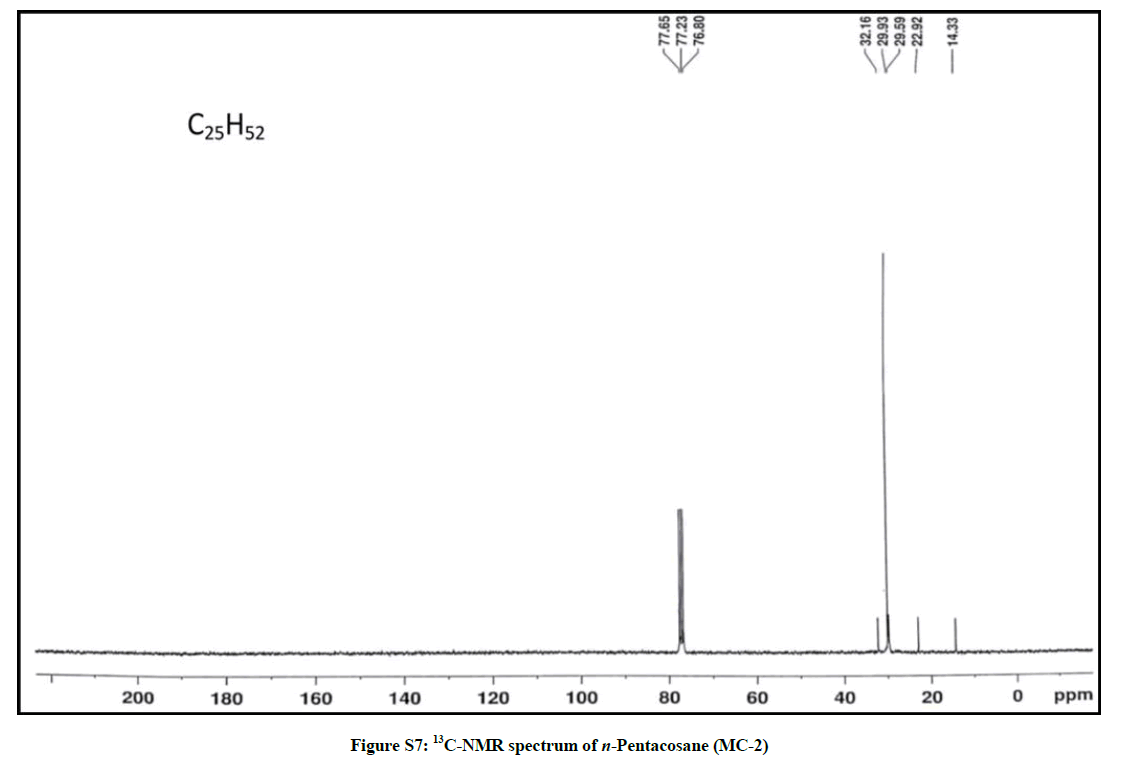
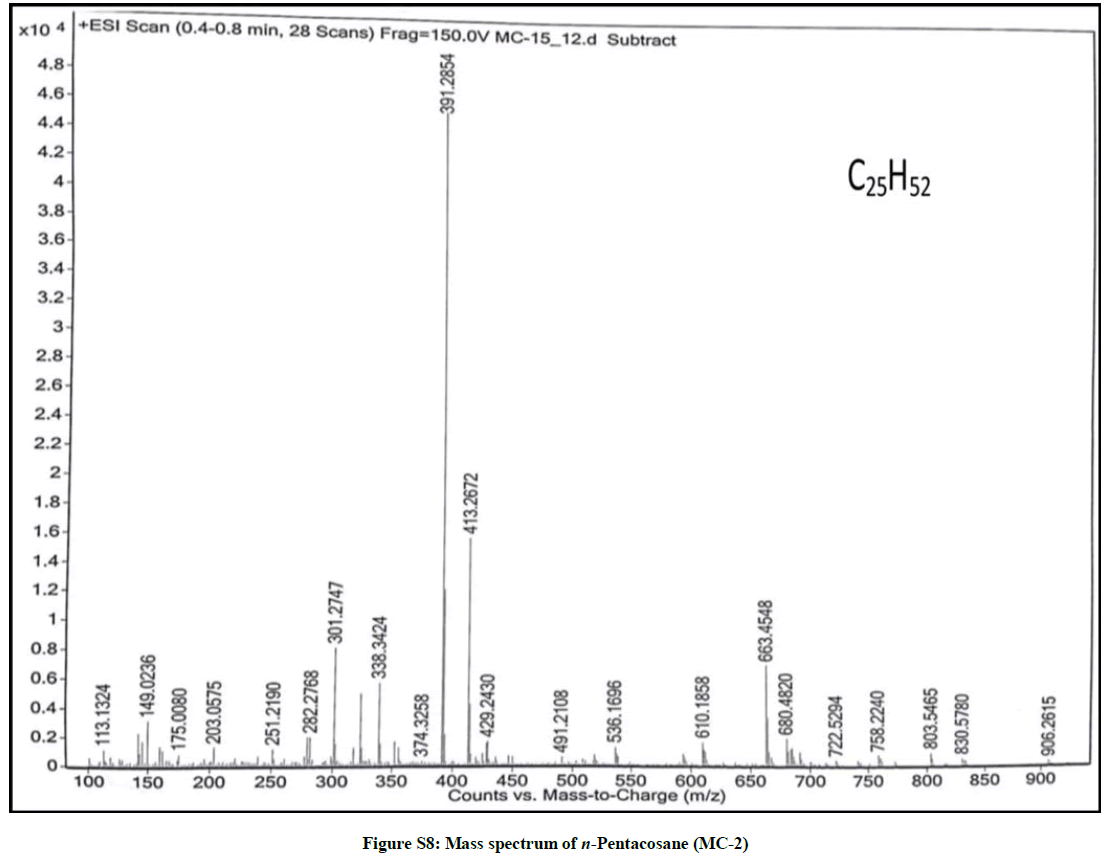
The IR spectrum of compound MC-2 displayed an absorption band for long chain hydrocarbon at 757 cm-1. The absence of any prominent band in the range 3500-3200 cm-1 and 1750-1600 cm-1 indicated the saturated nature of the molecule without any functional group. Its mass spectrum had a molecular ion peak at m/z 352 consistent with the molecular formula of a saturated hydrocarbon, C25H52. The absence of [M–Me]+ ion peak suggested straight nature of the molecule. The 1H-NMR spectrum of MC-2 showed two three-proton triplets at δ 0.89 (J= 6.0 Hz) and 0.85 (J= 7.2 Hz) assigned to the terminal C-1 and C-25 primary methyl protons, respectively. Six two - proton multiplets at δ 1.64, 1.50, 1.45, 1.41, 1.38 and 1.33 and two broad singlets at δ 1.29 (6H) and 1.25(28 H) were allied with the methylene protons. The 13C-NMR spectrum of MC-2 showed two signals for methyl carbons at δ 14.33 (C-1 and C-25) and between δ 22.92 to 32.16 for methylene carbons. The absence of any signal beyond δ 1.64 in the 1H-NMR spectrum and δ 32.16 in the 13C-NMR spectrum indicated the unsaturated nature of the molecule and absence of any functional group in it. The above mentioned spectral data interpretation led us to elucidate the structure of MC-2 as npentacosane (Figure 2).
Compound MC-3
Elution of the column with petroleum ether-chloroform (70: 30) furnished colorless crystals of MC-3, 81 mg (0.038% yield). Rf: 0.74 (benzene-chloroform- methanol, 2.5: 4.5: 0.6). Melting point: 81ºC- 82ºC; IR υmax (KBr): 2927, 2854, 1466, 1215, 1075, 929, 757 cm-1; 1H-NMR (CDCl3): δ (ppm)=1.44 (2H, m, CH2), 1.41 (2H, m, CH2), 1.33 (2H, m, CH2), 1.31 (2H, m, CH2), 1.30 (2H, m, CH2), 1.28 (8H, brs, 4 × CH2), 1.25 (48H, brs, 24 × CH2), 0.89 (3H, t, J=6.0 Hz, Me-1), 0.85 (3H, t, J=6.9 Hz, Me-35); 13C-NMR (CDCl3): δ (ppm)=32.15 (CH2), 30.43 (CH2), 29.92 (CH2), 29.58 (29 × CH2), 22.91 (CH2), 14.33 (Me-1, Me-35). ESI MS m/z (rel. int.): 492 [M]+(C35H72) (1.3).
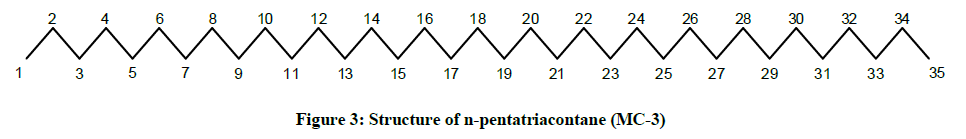
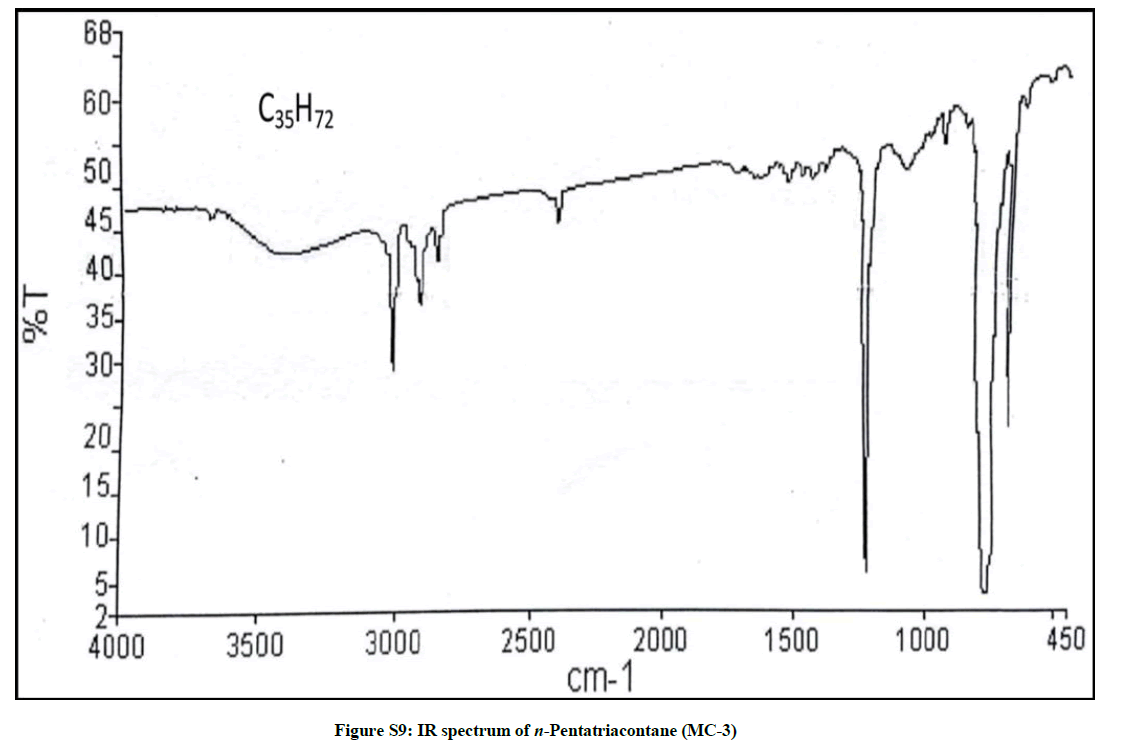
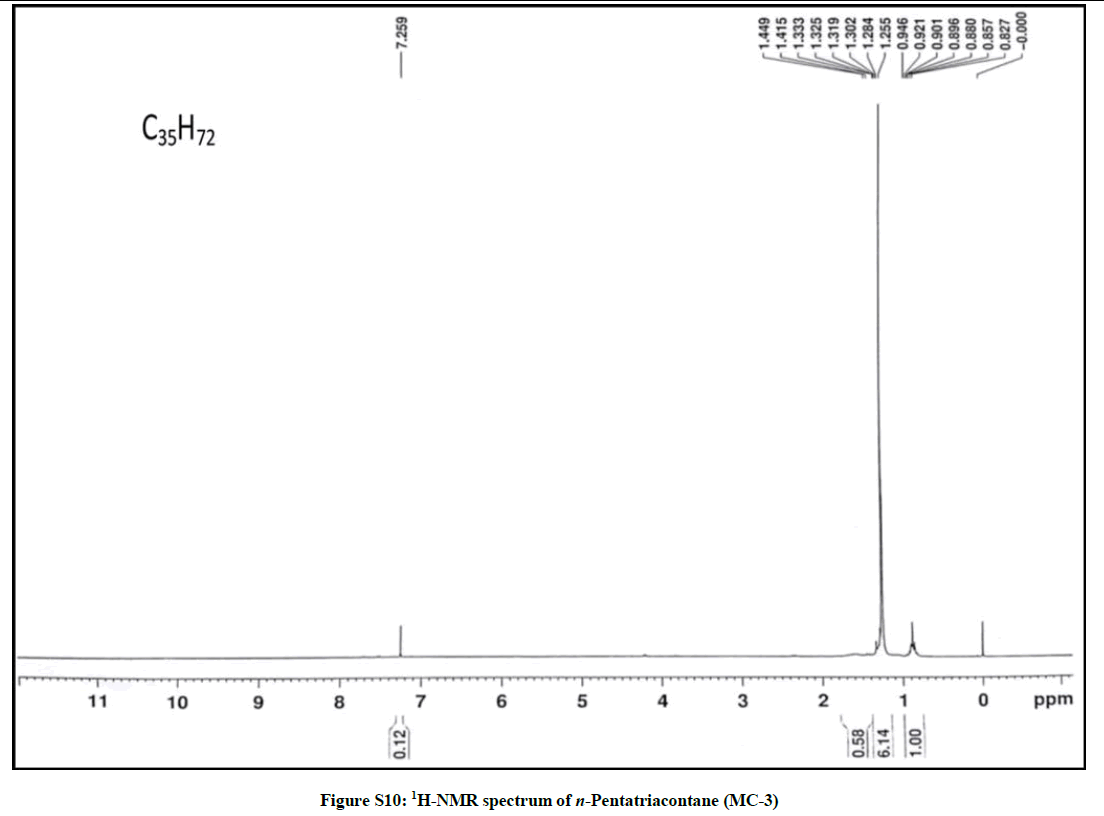
The IR spectrum of MC-3 demonstrated the presence of an absorption band for long chain hydrocarbon at 757 cm-1. The absence of any distinct band in the range 3500-3200 cm-1 and 1750-1600 cm-1 indicated the saturated nature of the molecule and ruled out the existence of any functional group. The mass spectrum had a molecular ion peak at m/z 492 corresponding to the molecular formula of a saturated hydrocarbon, C35H72. The deficiency of [M – Me]+ ion peak indicated the straight chain nature of the molecule. The 1H-NMR spectrum of MC-3 showed two three - proton triplets at δ 0.89 (J=6.0 Hz) and 0.85 (J=6.9 Hz) accounted to the terminal C-1 and C-35 primary methyl protons, respectively. Five two-proton multiplets from δ 1.44-1.30 and two broad singlets at δ 1.28 (8H) and 1.25 (48 H) were associated with the methylene protons. The 13C-NMR spectrum of MC-3 exhibited two signals at δ 14.33 due to presence of methyl groups at C-1 and C-35 and between δ 22.91 to 32.15 ascribed to the presence of methylene carbons. The absence of any signal beyond δ 1.44 in the 1H-NMR spectrum and δ 32.15 in the 13C-NMR spectrum supported the unsaturated nature of the molecule and absence of any functional group in it. This discussion of the spectral data analysis led us to explicate the structure of MC-3 as n-pentatriacontane (Figure 3).
Compound MC-4
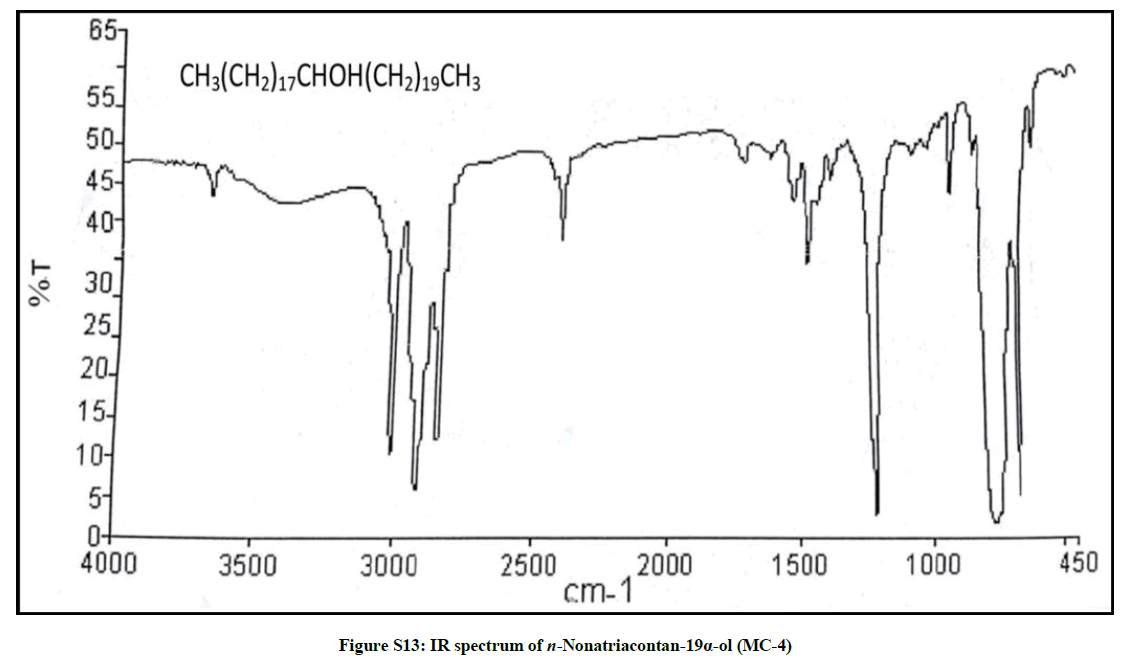
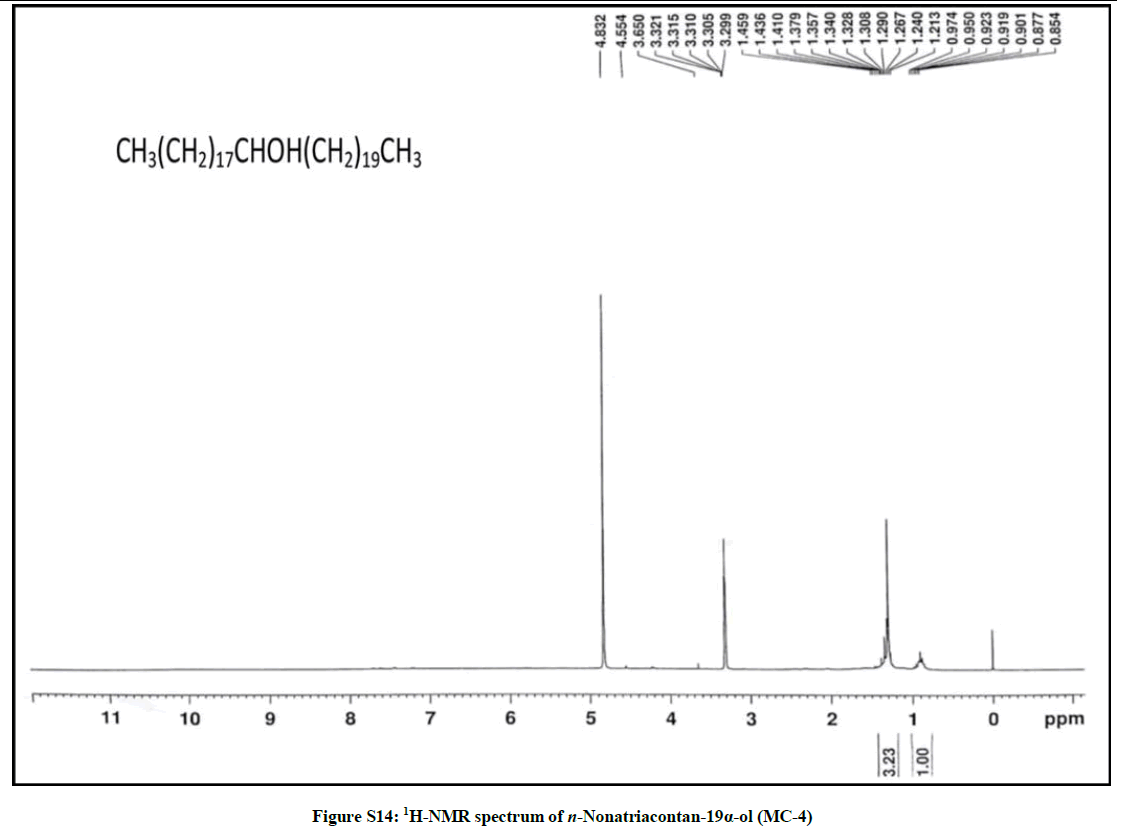
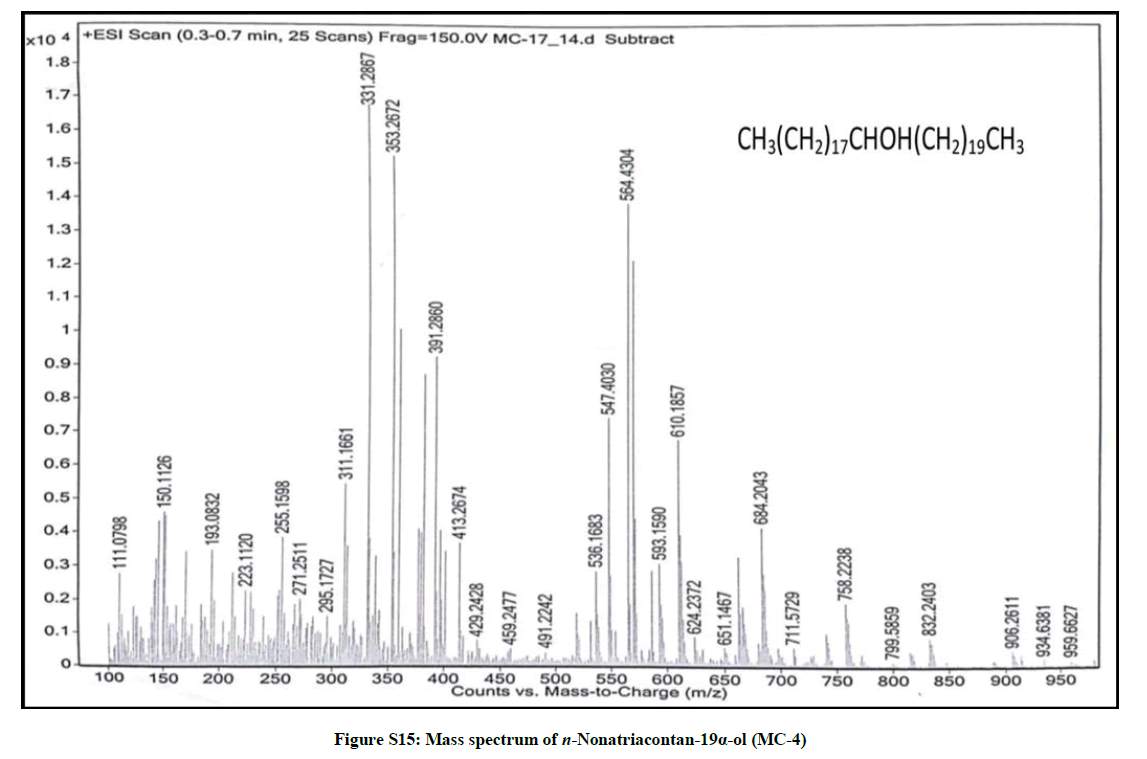
Elution of the column with chloroform-methanol (99.1: 0.1) produced colorless crystals of MC-4, 94 mg (0.044% yield). Rf: 0.55 (benzene-chloroform- methanol, 2.5: 4: 0.6). Melting point: 83ºC-84ºC; IR υmax (KBr): 3401, 2926, 2854, 1602, 1466, 1378, 1215, 1079, 1020, 929, 849, 757 cm-1; 1H-NMR (MeOD): δ (ppm)=3.31 (1H, m, w1/2=6.6 Hz, H-19β), 1.45 (2H, m, CH2), 1.43 (2H, m, CH2), 1.41 (2H, m, CH2), 1.37 (2H, m, CH2), 1.34 (2H, m, CH2), 1.32 (2H, m, CH2), 1.30 (2H, m, CH2), 1.26 (6H, s, 3 × CH2), 1.24 (2H, m, CH2), 0.92 (3H, t, J=8.1Hz, Me-1), 0.87 (3H, t, J=6.9Hz, Me-39); 13C-NMR (MeOD): δ (ppm)=73.27 (C-19), 45.31 (C-18), 41.16 (C-20), 34.11 (C-20), 30.89 (28 × CH2), 30.28 (CH2), 29.51 (CH2), 27.42 (CH2), 25.19 (CH2), 22.68 (CH2), 14.16 (Me-1), 14.07 (Me-39). ESI MS m/z (rel. int.): 564 [M]+(C39H80O) (68.5), 311 [CH3(CH2)19CHOH]+ (19.3), 253 [M-311, CH3(CH2)17]+ (12.4).
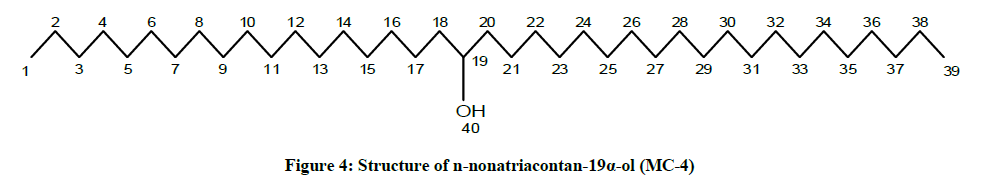
The compound MC-4 displayed IR absorption bands for a hydroxyl group (3401 cm-1) and long aliphatic chain (757 cm-1). Its mass spectrum exhibited a molecular ion peak at m/z 564 consequent to the molecular formula of a saturated aliphatic alcohol, C39H80O. The prominent ion peaks generating at m/z 311 [CH3 (CH2)10CHOH]+ and 253 [CH3(CH2)17] due to fission of linkage between C-18 and C-19 suggested the occurrence of the hydroxyl group at C-19. The 1H NMR spectrum of MC-4 displayed a one-proton multiplet at δ 3.31 with half width of 6.6 Hz associated with β-oriented carbinol H-19 proton. Two three - proton triplets at δ 0.92 (J=8.1 Hz) and 0.87 (J=6.9 Hz) were accounted to terminal C-1 and C-39 primary methyl protons respectively. Seven two-proton multiplets from δ 1.45-1.30 and two broad singlets at δ 1.26 (6H) and 1.24 (52 H) were ascribed to the remaining methylene protons. The 13C-NMR spectrum of MC-4 displayed the signals for carbinol carbon at δ 73.27 (C-19), methyl carbons at δ 14.16 (C-1) and 14.07 (C-39) and methylene carbons in the range of δ 45.31 to 22.68. The deficiency of any signal further δ 3.54 in 1H NMR and 66.11 in 13C-NMR suggested the saturated nature of the molecule. On the basis of the foregoing discussion the structure of the MC-4 has been formulated as n-nonatriacontan-19α-ol (Figure 4), an unknown aliphatic alcohol.
Compound MC-5
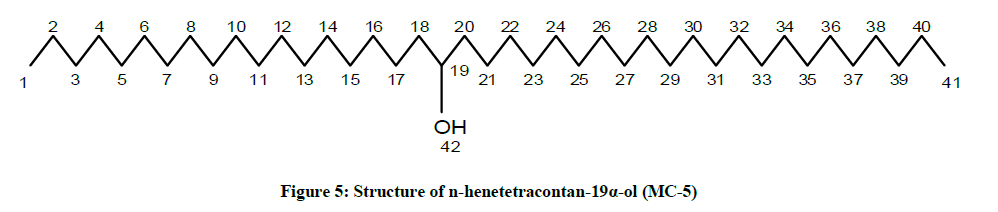
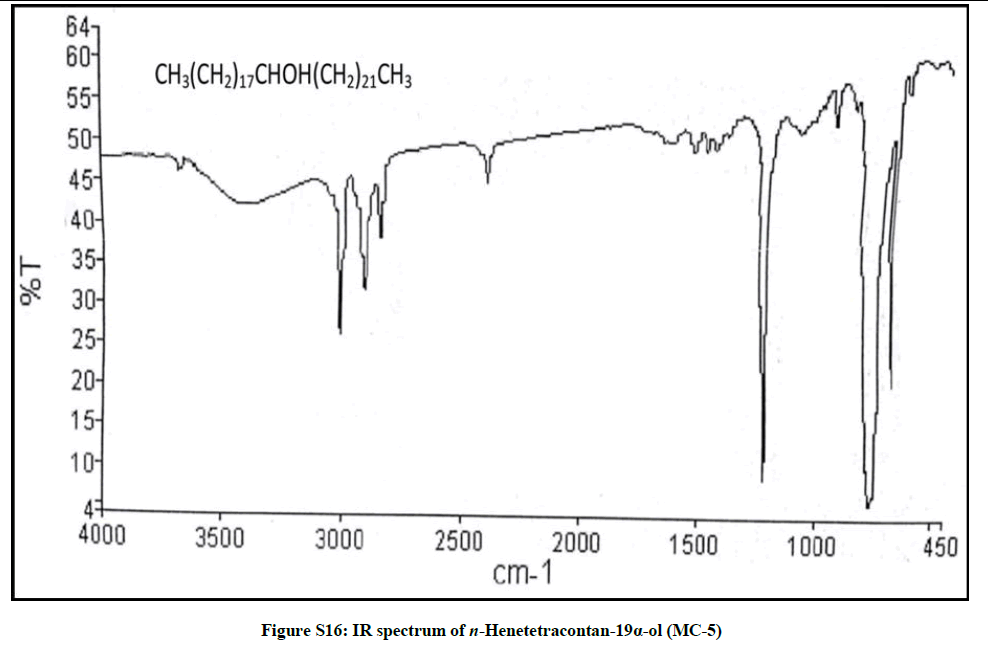
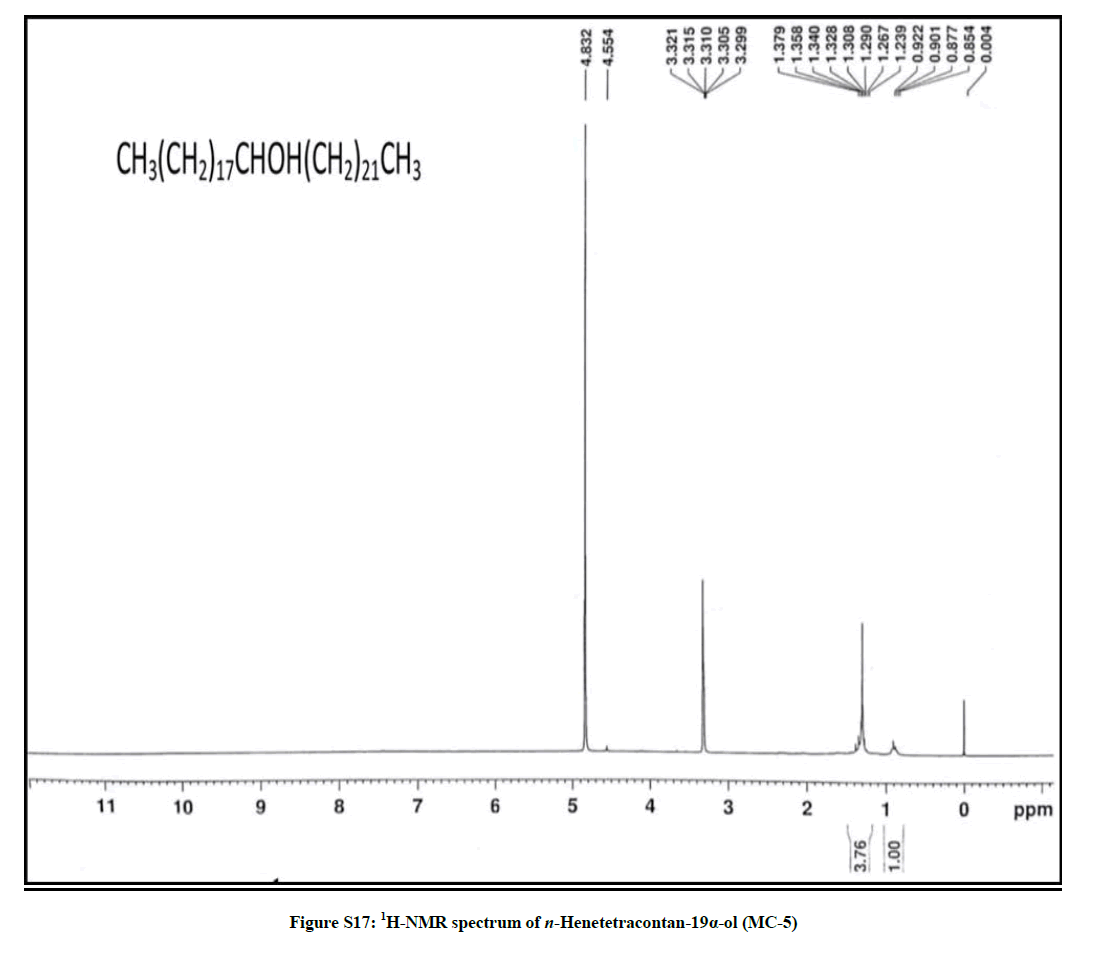
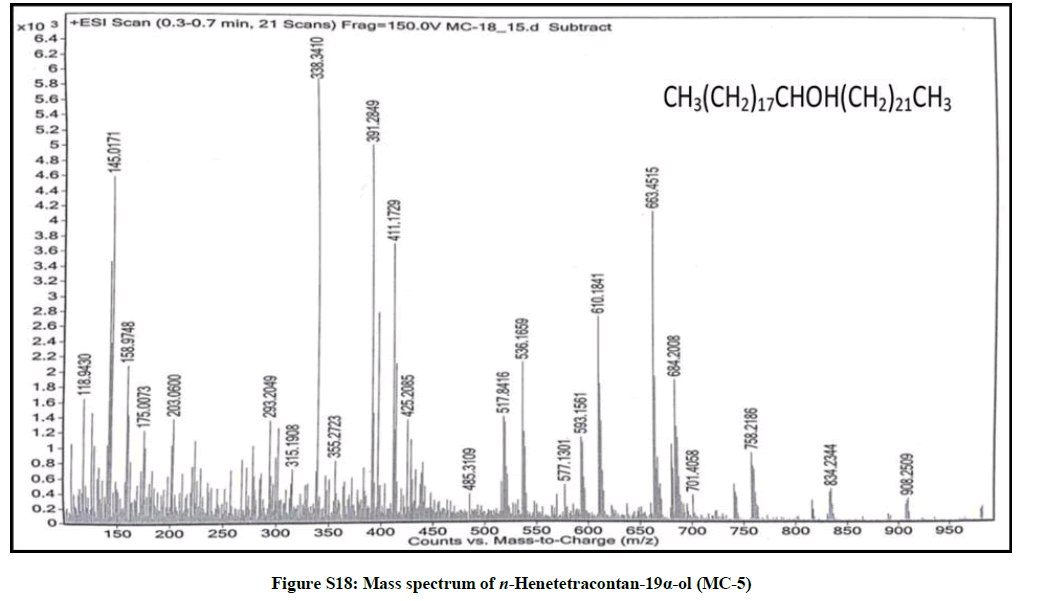
Elution of the column with chloroform-methanol (99.1: 0.1) gave a semisolid mass of MC-5, 79 mg (0.037% yield). Rf: 0.58 (Benzene-chloroform- methanol, 2.5: 4: 0.6). Melting point: 75ºC-76ºC. IR υmax (KBr): 3400, 2926, 2854, 1467, 1422, 1215, 1072, 929, 756 cm-1; 1H-NMR (MeOD): δ (ppm)=3.31 (1H, m, w1/2=6.6 Hz, H-19β), 1.37 (2H, m, CH2), 1.35 (2H, m, CH2), 1.34 (2H, m, CH2), 1.32 (2H, m, CH2), 1.30 (2H, m, CH2), 1.29 (4H, s, 2 × CH2), 1.26 (H, brs, CH2), 1.23 (2H, m, CH2), 0.90 (3H, t, J=6.3 Hz, Me-1), 0.87 (3H, t, J=6.9Hz, Me-41); 13C-NMR (MeOD): δ (ppm)=68.26 (C-19), 37.11 (CH2), 34.52 (CH2), 31.18 (CH2), 30.89 (CH2), 29.84 (CH2), 27.60 (CH2), 24.71 (CH2), 22.69 (CH2), 16.16 (C-1), 14.18 (C-41). ESI MS m/z (rel. int.): 592 [M]+(C41H84O) (18.7), 339 [CH3(CH2)21CHOH]+ (100), 253 [M-339, CH3(CH2)17]+(1.1)
The compound MC-5 exhibited characteristic IR absorption bands for a hydroxyl group (3400 cm-1) and long aliphatic chain (756 cm-1). Its mass spectrum displayed a molecular ion peak at m/z 592 corresponding to the molecular formula of a saturated aliphatic alcohol, C41H84O. The distinct ion peaks were produced at m/z 339 [CH3(CH2)21CHOH]+ and 253 [CH3(CH2)17] due to fission of the linkage between C-18 and C-19 suggesting the occurrence of hydroxyl group at C-19. The 1H-NMR spectrum of MC-5 showed a one-proton multiplet at δ 3.31 with half width of 6.6 Hz associated with β-oriented carbinol H-19 proton. Two three- proton triplets at δ 0.90 (J=6.3 Hz) and 0.87 (J=6.9 Hz) were ascribed to terminal C-1 and C-41 primary methyl protons, respectively. Six two-proton multiplets between δ 1.37-1.30 and at δ 1.23 and as broad singlets at δ 1.29 (4H) and 1.26 (30 H) were associated to the methylene protons. The 13C-NMR spectrum of MC-5 displayed the signals for carbinol carbon at δ 68.26 (C-19), methyl carbons at δ 16.16 (C-1) and 14.18 (C-41) and methylene carbons between δ 22.69 to 37.11. The deficiency of any signal ahead of δ 3.31 in 1H-NMR and δ 68.26 in 13C-NMR supported the saturated nature of the compound. On the basis of the spectral data analysis the structure of the compound MC-5 has been elucidated as n-henetetracontan-19α-ol (Figure 5), a new aliphatic alcohol.
Compound MC-6
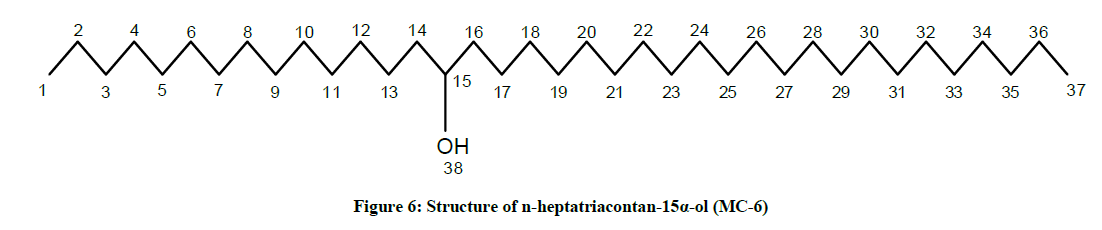
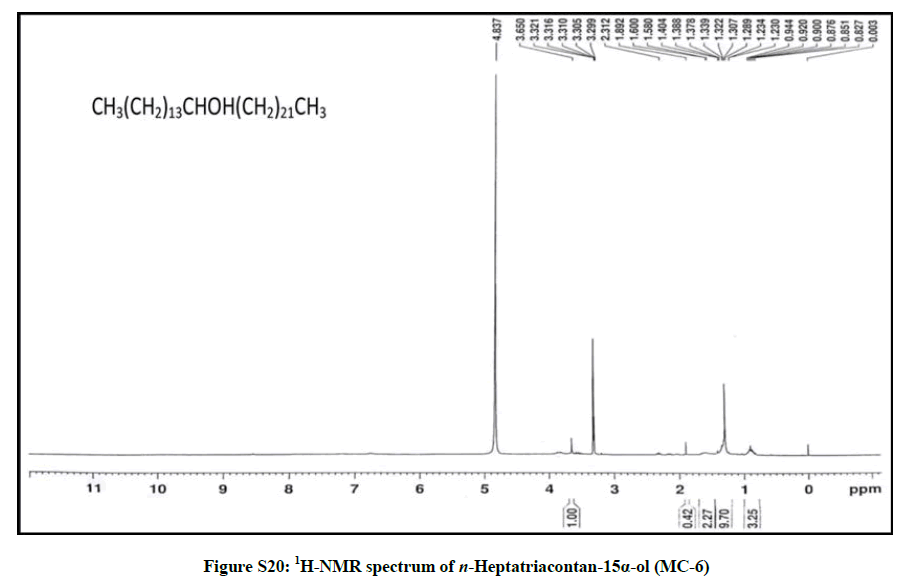
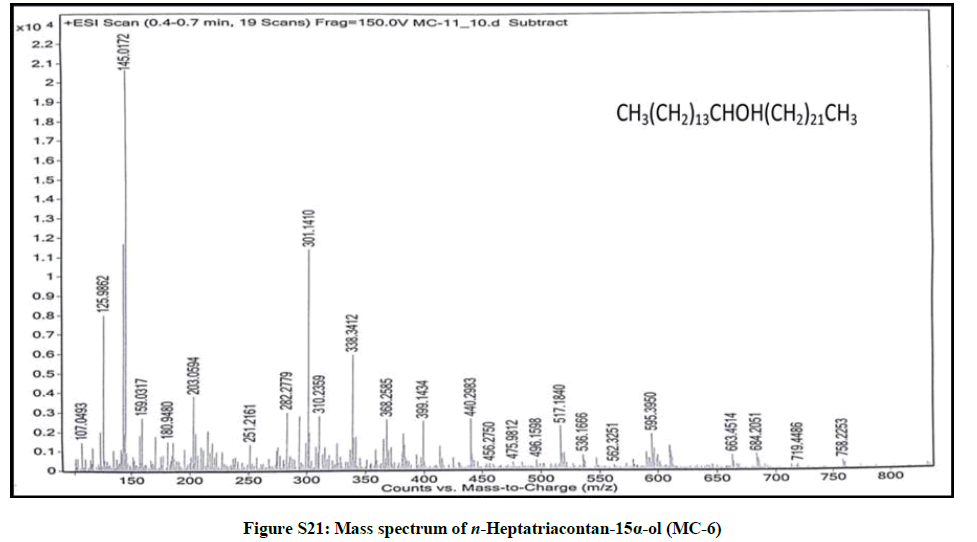
Elution of the column with chloroform-methanol (99.8: 0.2) gave colorless amorphous powder of MC-6, 73 mg (0.034% yield). Rf: 0.73 (chloroform-methanol, 10: 1). Melting point: 81ºC-82ºC; IR υmax (KBr): 3401, 2927, 2854, 1618, 1475, 1215, 1072, 929, 757 cm-1; 1H-NMR (MeOD): δ (ppm)=3.65 (1H, m, w1/2=6.6Hz, H-15β), 2.31 (2H, m, CH2), 1.89 (2H, m, CH2), 1.60 (2H, m, CH2), 1.58 (2H, m, CH2), 1.40 (2H, m, CH2), 1.38 (2H, m, CH2), 1.33 (2H, m, CH2), 1.32 (2H, m, CH2), 1.30 (2H, m, CH2), 1.28 (48 H, brs, 2H α CH2), 1.23 (2H, m, CH2), 0.92 (3H, t, J=6.0 Hz, Me-1), 0.85 (3H, t, J=7.2 Hz, Me-37); 13C-NMR (MeOD): δ (ppm)=68.21 (C-15), 45.65 (CH2), 37.23 (CH2), 34.48 (CH2), 30.89 (26 α CH2), 29.15 (CH2), 28.67 (CH2), 25.33 (CH2), 24.74 (CH2), 22.68 (CH2), 16.18 (C-1), 14.05 (C-37). ESI MS m/z (rel. int.): 536 [M]+ (C37H76O) (5.8), 339 [CH3(CH2)21CHOH]+(14.8).
The compound MC-6 showed IR absorption bands for hydroxyl group (3401 cm-1) and long aliphatic chain (757 cm-1). Its mass spectrum exhibited a molecular ion peak at m/z 536 corresponding to the molecular formula of a saturated aliphatic alcohol, C37H76O. The mass spectrum showed CnH2n and CnH2n-1 fragmentation peaks most of which were separated by 14 mass units indicating the straight chain characteristic of the compound. The prominent ion peak arising at m/z 339 [CH3 (CH2)21CHOH]+ due to fission between C-14 and C-15 indicated the presence of the hydroxyl group at C-15. The 1H-NMR spectrum of MC-6 showed a one-proton multiplet at δ 3.65 with half width of 6.6 Hz ascribed to β- oriented carbinol H-15 proton. Two three proton triplets at δ 0.92 (J=6.0 Hz) and 0.85 (J=7.2 Hz) were accounted to terminal C-1 and C-37 primary methyl protons, respectively. Nine two - proton multiplets at from δ 2.31 to 1.23 and a broad singlet at δ 1.28 (48 H) were associated with the methylene protons. The 13C-NMR spectrum of MC-6 exhibited signals for carbinol carbon at δ 68.21 (C-15), methyl carbons at δ 16.18 (C-1) and 14.05 (C-37) and methylene carbons between δ 45.65- 22.68. The absence of any signal ahead of δ 3.54 in 1H-NMR and 66.11 in 13C-NMR further supported that the molecule possessed the saturated nature. On the basis of these evidences the structure of MC-6 was elucidated as: n-heptatriacontan-15α-ol (Figure 6).
PASS prediction
The PASS computer program, which is able to simultaneously predict more than one thousand biological and toxicological activities from only the structural formulas of the chemicals, was used to predict the biological activity profile of the compounds isolated from Momordica charantia. Various novel pharmaceuticals have been discovered using PASS prediction. This application is a wonderful gift of technology to the mankind because it helps to discover new, potent and safe medicinal agents. It also provides an excellent opportunity for the multidisciplinary professionals (e.g. Pharma and Computational experts) to work together on a single platform and serve the society. Currently, PASS web-service is being utilised by more than 8700 registered users from more than 70 countries. Predictions for more than 250000 organic compounds have been obtained from this computer program. More than 4000 pharmacological effects, specific toxicities, mode of action, effect on gene expression, the interaction of metabolic enzymes, etc. have been predicted so far.
The predicted biological activities of the isolated phytoconstituents have been mentioned in Tables 1-6.
| Percentage activity | Percentage inactivity | Name of activity |
|---|---|---|
| 95.4 | 0.2 | Sugar-phosphatase inhibitor |
| 95 | 0.2 | Acrocylindropepsin inhibitor |
| 94 | 0.2 | Carboxypeptidase Taq inhibitor |
| 93.5 | 0.2 | Cutinase inhibitor |
| 93 | 0.2 | Pullulanase inhibitor |
| 92.6 | 0.3 | CYP2J substrate |
| 92.5 | 0.3 | Taurine dehydrogenase inhibitor |
| 92.4 | 0.4 | Phobic disorders treatment |
| 91.8 | 0.3 | CYP2J2 substrate |
| 91.5 | 0.2 | Poly(alpha-L-guluronate) lyase inhibitor |
| 91 | 0.2 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 90.7 | 0.3 | Dextranase inhibitor |
Table 1: Biological activity for n-Tetracosane
| Percentage activity | Percentage inactivity | Name of activity |
|---|---|---|
| 95.4 | 0.2 | Sugar-phosphatase inhibitor |
| 95 | 0.2 | Acrocylindropepsin inhibitor |
| 95 | 0.2 | Chymosin inhibitor |
| 95 | 0.2 | Saccharopepsin inhibitor |
| 94 | 0.2 | Carboxypeptidase Taq inhibitor |
| 93 | 0.2 | Pullulanase inhibitor |
| 92.9 | 0.6 | CYP2C12 substrate |
| 91 | 0.2 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 90.4 | 0.2 | Poly(beta-D-mannuronate) lyase inhibitor |
| 89 | 0.3 | Cl--transporting ATPase inhibitor |
| 89 | 0.3 | Lysine 2,3-aminomutase inhibitor |
Table 2: Biological activity for n-Pentacosane
| Percentage activity | Percentage inactivity | Name of activity |
|---|---|---|
| 95.4 | 0.2 | Sugar-phosphatase inhibitor |
| 95 | 0.2 | Acrocylindropepsin inhibitor |
| 95 | 0.2 | Chymosin inhibitor |
| 95 | 0.2 | Saccharopepsin inhibitor |
| 94 | 0.2 | Carboxypeptidase Taq inhibitor |
| 93.5 | 0.3 | Alkenylglycerophosphocholine hydrolase inhibitor |
| 93 | 0.2 | Pullulanase inhibitor |
| 91.8 | 0.3 | CYP2J2 substrate |
| 91.5 | 0.4 | Sphinganine kinase inhibitor |
| 90.7 | 0.3 | Dextranase inhibitor |
Table 3: Biological activity for n-Pentatriacontane
| Percentage activity | Percentage inactivity | Name of activity |
|---|---|---|
| 95.5 | 0.2 | Alkylacetylglycerophosphatase inhibitor |
| 95.3 | 0.2 | Acrocylindropepsin inhibitor |
| 95.3 | 0.2 | Chymosin inhibitor |
| 95.3 | 0.2 | Saccharopepsin inhibitor |
| 94.8 | 0.3 | Sphinganine kinase inhibitor |
| 94.4 | 0.2 | Sugar-phosphatase inhibitor |
| 94.3 | 0.2 | Acylcarnitine hydrolase inhibitor |
| 93.7 | 0.3 | Polyporopepsin inhibitor |
| 93 | 0.1 | Beta-mannosidase inhibitor |
| 92.9 | 0.6 | CYP2C12 substrate |
| 92.1 | 0.3 | Fucosterol-epoxide lyase inhibitor |
| 91.9 | 0.4 | Alkenylglycerophosphocholine hydrolase inhibitor |
Table 4: Biological Activity for n-Nonatriacontan-19α-ol
| Percentage activity | Percentage inactivity | Name of activity |
|---|---|---|
| 95.5 | 0.2 | Alkylacetylglycerophosphatase inhibitor |
| 95.3 | 0.2 | Acrocylindropepsin inhibitor |
| 95.3 | 0.2 | Chymosin inhibitor |
| 95.3 | 0.2 | Saccharopepsin inhibitor |
| 94.8 | 0.3 | Sphinganine kinase inhibitor |
| 94.4 | 0.2 | Sugar-phosphatase inhibitor |
| 94.3 | 0.2 | Acylcarnitine hydrolase inhibitor |
| 93.7 | 0.3 | Polyporopepsin inhibitor |
| 93.6 | 0.4 | 5 Hydroxytryptamine release stimulant |
| 93.4 | 0.1 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 93 | 0.1 | Beta-mannosidase inhibitor |
Table 5: Biological activity for n-Henetetracontan-19α-ol
| Percentage activity | Percentage inactivity | Name of activity |
|---|---|---|
| 95.5 | 0.2 | Alkylacetylglycerophosphatase inhibitor |
| 95.3 | 0.2 | Acrocylindropepsin inhibitor |
| 95.3 | 0.2 | Chymosin inhibitor |
| 95.3 | 0.2 | Saccharopepsin inhibitor |
| 94.8 | 0.3 | Sphinganine kinase inhibitor |
| 94.4 | 0.2 | Sugar-phosphatase inhibitor |
| 94.3 | 0.2 | Acylcarnitine hydrolase inhibitor |
| 93.7 | 0.3 | Polyporopepsin inhibitor |
| 93.6 | 0.4 | 5 Hydroxytryptamine release stimulant |
| 93.4 | 0.1 | Xylan endo-1,3-beta-xylosidase inhibitor |
Table 6: Biological activity for n-Heptatriacontan-15α-ol
Conclusion
Two unknown aliphatic alcohols n-nonatriacontan-19α-ol and n-henetetracontan-19α-ol along with four known phytomolecules: n-tetracosane, n-pentacosane, n-pentatriacontane and n-heptatriacontan-15α-ol were isolated from the ethanolic extract of the seeds of Momordica charantia Linn. The various chromatographic techniques might use these compounds as fingerprinting markers. Thus these shall be beneficial for establishing the identity, quality and purity of the drug. These phytoconstituents shall be explored for their pharmacological activity, especially the new compounds. For this the PASS prediction for the biological activity spectra shall be of great help. The Biological Activity Spectrum of n-tetracosane, n-pentacosane and n-pentatriacontane exhibited their maximun probability to be active as Sugar-phosphatase inhibitor, Acrocylindropepsin inhibitor, Chymosin inhibitor and Saccharopepsin inhibitor while that of n- nonatriacontan-19α-ol, henetetracontan-19α-ol and n-heptatriacontan-15α-ol showed their highest probability to be potent as Alkylacetylglycerophosphatase inhibitor,Acrocylindropepsin inhibitor, Chymosin inhibitor, Saccharopepsin inhibitor, Sphinganine kinase inhibitor. Thus, the PASS prediction of the various isolated compounds also supported the fact that the compounds having similar chemical structures possess similar Biological activity spectrum.
Acknowledgement
The authors are thankful to the Principal, KIET School of Pharmacy for motivation and to the librarians of the KIET library and NISCAIR library, New Delhi for providing the literature.
References
- X. Xua, B. Shanb, C.H. Liaob, J.H. Xiea, P.W. Wena, J.Y. Shida, Int. J. Biol. Macromol., 2015, 81, 538-543.
- B. Svobodova, L. Barrosa, R.C. Calhelhaa, S. Helenoa, M.J. Alvesd, S. Walcotte, Ind. Crops Prod., 2016.
- S. Kumar, J. Ashish, N. Satish, I.J.B.R., 2011, 2(11), 579-587.
- D. Katiyar, V. Singh, M. Ali, Biochemistry and Therapeutic Uses of Medicinal Plants, Discovery Publishing House Pvt. Ltd., New Delhi (India), 2017, 1-33.
- S.J. Yang, J.M. Choi, S.E. Park, E.J. Rhee, W.Y. Lee, K.W. Oh, J. Nutr. Biochem., 2014.
- C. Day, T. Cartwright, J. Provost, C.J. Bailey, Planta Med., 1990, 56, 426-429.
- A. Raman, C. Lau, Phytomedicine., 1996, 2(4), 349-362.
- B. Ajitha, Y.A.K. Reddy, S. Reddy, J. Photochem. Photobiol., 2015.
- J. Husain, I.J. Tickle, S.P. Wood, F.E.B.S. letters, 1994, 342, 154-158.
- H. Xie,S. Huang, H. Deng, Z. Wu, A. Ji, J. Chinese Med. Material., 1998, 21, 458-459.
- Y.R. Yuan, Y.N. He, J.P. Xiong, Z.X. Xia, ActaCrystallogra Sec D-BiolCrystallogra., 2011, 55, 1144-1151.
- A Parkash,T.B. Ng, W.W. Tso, J. Pept. Res., 2002, 59, 197-202.
- D. Katiyar, V. Singh, M. Ali, Int. J. Res. Ayurveda. Pharm., 2016, 7(3), 169-173.
- J. Pitiphanpong, S. Chitprasert, M. Goto, W. Jiratchariyakul, M. Sasaki, A. Shotipruk A, Purif. Technol., 2007, 52, 416-422.
- A. Dans, M. Villarruz, C. Jimeno, M. Javelosa, J. Chua, R. Bautista, G.G. Velez, J. Clin. Epidemiol., 2007, 60(6), 554-559.
- M. Sasa, I. Inoue, Y. Shinoda, S. Takahashi, M. Seo, T. Komoda, T. Awata, S. Katayama, J. Atheroscler. Thromb., 2009, 16(6), 888-892.
- G. Handa, J. Singh, M.L. Sharma, A. Kaul, R. Zafar, Indian J. Nat. Prod., 1990, 6, 16-19.
- A. Kochhar and M. Nagi, J. Med. Food., 2011, 8(4), 545-549.
Supplementary Material
Spectrums of isolated phytoconstituents (Figures S1-S21)
Tables for prediction of biological activity (Tables S1- S6)
| Percentage Activity | Percentage Inactivity | Name of Activity |
|---|---|---|
| 95.4 | 0.2 | Sugar-phosphatase inhibitor |
| 95 | 0.2 | Acrocylindropepsin inhibitor |
| 95 | 0.2 | Chymosin inhibitor |
| 95 | 0.2 | Saccharopepsin inhibitor |
| 94.2 | 0.2 | Acylcarnitine hydrolase inhibitor |
| 94.1 | 0.2 | Alkylacetylglycerophosphatase inhibitor |
| 94 | 0.2 | CarboxypeptidaseTaq inhibitor |
| 93.7 | 0.2 | IgA-specific serine endopeptidase inhibitor |
| 93.5 | 0.2 | Cutinase inhibitor |
| 93 | 0.2 | Pullulanase inhibitor |
| 92.6 | 0.3 | CYP2J substrate |
| 92.5 | 0.3 | Taurine dehydrogenase inhibitor |
| 92.9 | 0.6 | CYP2C12 substrate |
| 92.4 | 0.4 | Phobic disorders treatment |
| 91.8 | 0.3 | CYP2J2 substrate |
| 91.5 | 0.2 | Poly(alpha-L-guluronate) lyase inhibitor |
| 91.3 | 0.2 | Exoribonuclease II inhibitor |
| 91 | 0.2 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 90.7 | 0.3 | Dextranase inhibitor |
| 90.4 | 0.2 | Poly(beta-D-mannuronate) lyase inhibitor |
| 89.5 | 0.5 | Antieczematic |
| 89.2 | 0.3 | Sarcosine oxidase inhibitor |
| 88.8 | 0.2 | Anthranilate-CoA ligase inhibitor |
| 88.1 | 0.4 | Glutamylendopeptidase II inhibitor |
| 87 | 0.4 | 5-O-(4-coumaroyl)-D-quinate 3'-monooxygenase inhibitor |
| 86 | 0.4 | Phosphatidylcholine-retinol O-acyltransferase inhibitor |
| 86.4 | 0.9 | Chlordeconereductase inhibitor |
| 86.9 | 1.9 | Membrane integrity agonist |
| 85.7 | 0.9 | Antiseborrheic |
| 85.1 | 0.3 | Electron-transferring-flavoprotein dehydrogenase inhibitor |
| 85.2 | 0.4 | 2-Hydroxymuconate-semialdehyde hydrolase inhibitor |
| 85 | 0.5 | Dehydro-L-gulonate decarboxylase inhibitor |
| 85.3 | 0.8 | Beta-adrenergic receptor kinase inhibitor |
| 84.5 | 0.9 | Protein-glutamate methylesterase inhibitor |
| 84 | 0.4 | Creatininase inhibitor |
| 83.9 | 0.5 | Fragilysin inhibitor |
| 83.8 | 0.4 | UDP-N-acetylglucosamine 4-epimerase inhibitor |
| 83.7 | 0.8 | Feruloyl esterase inhibitor |
| 82.9 | 0.5 | Omptin inhibitor |
| 82.3 | 0.2 | Oryzin inhibitor |
| 82.7 | 0.7 | Nicotinic alpha6beta3beta4alpha5 receptor antagonist |
| 82.9 | 0.9 | NADPH peroxidase inhibitor |
| 82.3 | 0.3 | Leukopoiesis stimulant |
| 82.7 | 0.9 | Glucose oxidase inhibitor |
| 81.5 | 0.5 | Vasoprotector |
| 81 | 0.6 | Glutathione thiolesterase inhibitor |
| 81.5 | 1.2 | Mucositis treatment |
| 80.2 | 0.6 | Pseudolysin inhibitor |
| 79.5 | 0.3 | Licheninase inhibitor |
| 79.4 | 0.2 | Dolichyl-diphosphooligosaccharide-protein glycotransferase inhibitor |
| 80.1 | 1 | Nicotinic alpha2beta2 receptor antagonist |
| 79.2 | 0.5 | N-benzyloxycarbonylglycine hydrolase inhibitor |
| 79.1 | 0.4 | NADH kinase inhibitor |
| 79 | 0.4 | Coccolysin inhibitor |
| 78.9 | 0.3 | Polyneuridine-aldehyde esterase inhibitor |
| 78.9 | 0.3 | Oxygen scavenger |
| 78.5 | 0.5 | L-glutamate oxidase inhibitor |
| 78 | 0.6 | 2-Hydroxyquinoline 8-monooxygenase inhibitor |
| 78 | 1.2 | Arylacetonitrilase inhibitor |
| 77 | 0.4 | Laccase inhibitor |
| 76.5 | 1.2 | Prostaglandin-E2 9-reductase inhibitor |
| 75.5 | 0.4 | Kidney function stimulant |
| 75 | 0.3 | Glycolate dehydrogenase inhibitor |
| 74.4 | 0.3 | 2-Hydroxy-3-oxoadipate synthase inhibitor |
| 74.2 | 0.5 | Sulfite reductase inhibitor |
| 73.1 | 0.7 | Hydrogen dehydrogenase inhibitor |
| 72.9 | 1.4 | Peptidyl-dipeptidaseDcp inhibitor |
| 71 | 0.8 | Histidine N-acetyltransferase inhibitor |
| 70 | 0.2 | N-methyl-2-oxoglutaramate hydrolase inhibitor |
Table S1: Biological Activity for n-tetracosane
| Percentage Activity | Percentage Inactivity | Name of Activity |
|---|---|---|
| 95.4 | 0.2 | Sugar-phosphatase inhibitor |
| 95 | 0.2 | Acrocylindropepsin inhibitor |
| 95 | 0.2 | Chymosin inhibitor |
| 95 | 0.2 | Saccharopepsin inhibitor |
| 94 | 0.2 | CarboxypeptidaseTaq inhibitor |
| 93 | 0.2 | Pullulanase inhibitor |
| 92.9 | 0.6 | CYP2C12 substrate |
| 91 | 0.2 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 90.4 | 0.2 | Poly(beta-D-mannuronate) lyase inhibitor |
| 89 | 0.3 | Cl--transporting ATPase inhibitor |
| 89 | 0.3 | Lysine 2,3-aminomutase inhibitor |
| 88.8 | 0.2 | Anthranilate-CoA ligase inhibitor |
| 87 | 0.3 | Thioredoxin inhibitor |
| 86.9 | 0.4 | Complement factor D inhibitor |
| 86.7 | 0.3 | Polyamine-transporting ATPase inhibitor |
| 86.8 | 0.4 | Cardiovascular analeptic |
| 86.7 | 0.4 | Ribulose-phosphate 3-epimerase inhibitor |
| 86.6 | 0.4 | Nitrate reductase (cytochrome) inhibitor |
| 86.4 | 0.3 | Amine dehydrogenase inhibitor |
| 86.3 | 0.3 | Phosphatidylglycerophosphatase inhibitor |
| 86.3 | 0.3 | Levanase inhibitor |
| 86.4 | 0.4 | GST A substrate |
| 86.2 | 0.4 | Venombin AB inhibitor |
| 86.4 | 0.9 | Chlordeconereductase inhibitor |
| 85.8 | 0.4 | Fusarinine-C ornithinesterase inhibitor |
| 85.5 | 0.3 | Trimethylamine-oxide aldolase inhibitor |
| 85.3 | 0.2 | Endopeptidase So inhibitor |
| 85.3 | 0.3 | Urethanase inhibitor |
| 85.8 | 0.8 | Mucomembranous protector |
| 86.9 | 1.9 | Membrane integrity agonist |
| 85.7 | 0.9 | Antiseborrheic |
| 85.1 | 0.3 | Electron-transferring-flavoprotein dehydrogenase inhibitor |
| 85.2 | 0.4 | 2-Hydroxymuconate-semialdehyde hydrolase inhibitor |
| 85 | 0.5 | Dehydro-L-gulonate decarboxylase inhibitor |
| 85.3 | 0.8 | Beta-adrenergic receptor kinase inhibitor |
| 85.3 | 0.8 | G-protein-coupled receptor kinase inhibitor |
| 84.8 | 0.4 | Dimethylargininase inhibitor |
| 85 | 0.6 | Glycosylphosphatidylinositol phospholipase D inhibitor |
| 84.6 | 0.3 | Rhamnulose-1-phosphate aldolase inhibitor |
| 84.7 | 0.4 | Glyceryl-ether monooxygenase inhibitor |
| 84.6 | 0.2 | Prostaglandin-A1 DELTA-isomerase inhibitor |
| 84.8 | 0.5 | Glucan endo-1,6-beta-glucosidase inhibitor |
| 84.6 | 0.3 | Methylamine-glutamate N-methyltransferase inhibitor |
| 84.4 | 0.2 | BRAF expression inhibitor |
| 84 | 0.2 | 2-Haloacid dehalogenase inhibitor |
| 84.5 | 0.9 | Protein-glutamate methylesterase inhibitor |
| 83.2 | 1.4 | Methylenetetrahydrofolatereductase (NADPH) inhibitor |
| 81.9 | 0.3 | 2-Haloacid dehalogenase (configuration-inverting) inhibitor |
| 81 | 0.6 | Glutathione thiolesterase inhibitor |
| 81.5 | 1.2 | Mucositis treatment |
| 80.4 | 0.5 | Lipoprotein lipase inhibitor |
| 80 | 0.3 | Procollagen N-endopeptidase inhibitor |
| 80.1 | 1 | Nicotinic alpha2beta2 receptor antagonist |
| 79 | 0.4 | Coccolysin inhibitor |
| 78.9 | 0.3 | Polyneuridine-aldehyde esterase inhibitor |
| 78.9 | 0.3 | Oxygen scavenger |
| 79 | 0.4 | 1, 4-Lactonase inhibitor |
| 79 | 0.5 | Limulus clotting factor C inhibitor |
| 78 | 1.2 | Arylacetonitrilase inhibitor |
| 77 | 0.4 | Laccase inhibitor |
| 76 | 0.4 | Tpr proteinase (Porphyromonasgingivalis) inhibitor |
| 76.1 | 0.7 | Phthalate 4, 5-dioxygenase inhibitor |
| 76.5 | 1.2 | Prostaglandin-E2 9-reductase inhibitor |
| 75 | 0.3 | Glycolate dehydrogenase inhibitor |
| 75.1 | 0.4 | Opheline kinase inhibitor |
| 75.1 | 0.4 | Taurocyamine kinase inhibitor |
| 75.3 | 2.7 | Antineurotic |
| 73 | 0.4 | Carbon-monoxide dehydrogenase inhibitor |
| 72.5 | 0.3 | Plastoquinol-plastocyaninreductase inhibitor |
| 72.1 | 1.5 | Aldehyde oxidase inhibitor |
| 71 | 0.8 | Histidine N-acetyltransferase inhibitor |
| 70 | 0.2 | N-methyl-2-oxoglutaramate hydrolase inhibitor |
Table S2: Biological Activity for n-pentacosane
| Percentage Activity | Percentage Inactivity | Name of Activity |
|---|---|---|
| 95.4 | 0.2 | Sugar-phosphatase inhibitor |
| 95 | 0.2 | Acrocylindropepsin inhibitor |
| 95 | 0.2 | Chymosin inhibitor |
| 95 | 0.2 | Saccharopepsin inhibitor |
| 94 | 0.2 | CarboxypeptidaseTaq inhibitor |
| 93.5 | 0.3 | Alkenylglycerophosphocholine hydrolase inhibitor |
| 93 | 0.2 | Pullulanase inhibitor |
| 91.8 | 0.3 | CYP2J2 substrate |
| 91.5 | 0.4 | Sphinganine kinase inhibitor |
| 91 | 0.2 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 90.7 | 0.3 | Dextranase inhibitor |
| 90.4 | 0.2 | Poly(beta-D-mannuronate) lyase inhibitor |
| 8.9 | 0.3 | Lysine 2,3-aminomutase inhibitor |
| 88.8 | 0.2 | Anthranilate-CoA ligase inhibitor |
| 87 | 0.4 | 5-O-(4-coumaroyl)-D-quinate 3'-monooxygenase inhibitor |
| 86.9 | 1.9 | Membrane integrity agonist |
| 85 | 0.6 | Glycosylphosphatidylinositol phospholipase D inhibitor |
| 84.5 | 0.9 | Protein-glutamate methylesterase inhibitor |
| 83.5 | 0.2 | Hydroxylamine oxidase inhibitor |
| 83.5 | 0.6 | Membrane permeability inhibitor |
| 83.2 | 0.3 | Beta-mannosidase inhibitor |
| 83.7 | 0.8 | Feruloyl esterase inhibitor |
| 82.9 | 0.5 | Omptin inhibitor |
| 82.8 | 0.5 | Bisphosphoglycerate phosphatase inhibitor |
| 82.5 | 0.2 | Phenylacetate-CoA ligase inhibitor |
| 82.7 | 0.4 | 3-Hydroxybenzoate 6-monooxygenase inhibitor |
| 82.3 | 0.3 | Leukopoiesis stimulant |
| 82.3 | 0.5 | NADPH-cytochrome-c2 reductase inhibitor |
| 81 | 0.4 | Lysostaphin inhibitor |
| 80 | 0.3 | Procollagen N-endopeptidase inhibitor |
| 80.2 | 0.6 | Pseudolysin inhibitor |
| 79.2 | 0.5 | N-benzyloxycarbonylglycine hydrolase inhibitor |
| 79.1 | 0.4 | NADH kinase inhibitor |
| 79 | 0.4 | 1,4-Lactonase inhibitor |
| 79 | 0.5 | Limulus clotting factor C inhibitor |
| 78 | 1.2 | Arylacetonitrilase inhibitor |
| 77.2 | 0.4 | Carminative |
| 77 | 0.4 | Laccase inhibitor |
| 76.1 | 0.7 | Phthalate 4,5-dioxygenase inhibitor |
| 76.5 | 1.2 | Prostaglandin-E2 9-reductase inhibitor |
| 75 | 0.3 | Glycolate dehydrogenase inhibitor |
| 75.1 | 0.4 | Opheline kinase inhibitor |
| 75.2 | 1 | ADP-thymidine kinase inhibitor |
| 74.4 | 0.4 | Di-trans,poly-cis-decaprenylcistransferase inhibitor |
| 74.4 | 0.4 | Adenomatous polyposis treatment |
| 73 | 0.5 | Tryptophanamidase inhibitor |
| 72.9 | 1.4 | Peptidyl-dipeptidaseDcp inhibitor |
| 72 | 1.4 | 2-Dehydropantoate 2-reductase inhibitor |
| 71 | 0.4 | Acylaminoacyl-peptidase inhibitor |
| 72.1 | 1.5 | Aldehyde oxidase inhibitor |
| 71 | 0.8 | Histidine N-acetyltransferase inhibitor |
| 70 | 0.2 | N-methyl-2-oxoglutaramate hydrolase inhibitor |
Table S3: Biological Activity for n-pentatriacontane
| Percentage Activity | Percentage Inactivity | Name of Activity |
|---|---|---|
| 95.5 | 0.2 | Alkylacetylglycerophosphatase inhibitor |
| 95.3 | 0.2 | Acrocylindropepsin inhibitor |
| 95.3 | 0.2 | Chymosin inhibitor |
| 95.3 | 0.2 | Saccharopepsin inhibitor |
| 94.8 | 0.3 | Sphinganine kinase inhibitor |
| 94.4 | 0.2 | Sugar-phosphatase inhibitor |
| 94.3 | 0.2 | Acylcarnitine hydrolase inhibitor |
| 93.7 | 0.3 | Polyporopepsin inhibitor |
| 93 | 0.1 | Beta-mannosidase inhibitor |
| 92.9 | 0.6 | CYP2C12 substrate |
| 92.1 | 0.3 | Fucosterol-epoxide lyase inhibitor |
| 91.9 | 0.4 | Alkenylglycerophosphocholine hydrolase inhibitor |
| 91.1 | 0.2 | Glucan 1,4-alpha-maltotriohydrolase inhibitor |
| 91 | 0.3 | CarboxypeptidaseTaq inhibitor |
| 90.1 | 0.4 | CYP2J2 substrate |
| 90 | 0.3 | Cutinase inhibitor |
| 89.9 | 0.3 | IgA-specific metalloendopeptidase inhibitor |
| 87.1 | 0.5 | Feruloyl esterase inhibitor |
| 86.6 | 0.2 | Alcohol dehydrogenase (acceptor) inhibitor |
| 86.4 | 0.3 | Trimethylamine-oxide aldolase inhibitor |
| 86.5 | 0.3 | All-trans-retinyl-palmitate hydrolase inhibitor |
| 86 | 0.2 | Anthranilate-CoA ligase inhibitor |
| 86.1 | 0.3 | Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase inhibitor |
| 85.9 | 0.3 | Poly(beta-D-mannuronate) lyase inhibitor |
| 85.1 | 0.4 | Vasoprotector |
| 84.9 | 0.2 | CYP4A11 substrate |
| 85.3 | 0.7 | Prostaglandin-E2 9-reductase inhibitor |
| 84.7 | 0.1 | Sclerosant |
| 84 | 0.4 | Venombin AB inhibitor |
| 83 | 0.3 | Endopeptidase So inhibitor |
| 82.1 | 0.4 | Allyl-alcohol dehydrogenase inhibitor |
| 81.5 | 0.8 | Membrane permeability inhibitor |
| 81 | 0.3 | Alkylglycerone-phosphate synthase inhibitor |
| 80.7 | 0.2 | Oryzin inhibitor |
| 80.1 | 0.8 | 5-O-(4-coumaroyl)-D-quinate 3'-monooxygenase inhibitor |
| 79.4 | 0.4 | Lactase inhibitor |
| 79.6 | 0.7 | Superoxide dismutase inhibitor |
| 79 | 0.3 | Aspergillopepsin I inhibitor |
| 79.1 | 0.5 | Polyamine-transporting ATPase inhibitor |
| 78.4 | 0.2 | Sphinganine-1-phosphate aldolase inhibitor |
| 80.1 | 2 | Chlordeconereductase inhibitor |
| 79 | 2 | Methylenetetrahydrofolatereductase (NADPH) inhibitor |
| 78 | 2.3 | Antieczematic |
| 77.4 | 2.4 | Antiseborrheic |
| 76 | 0.9 | Bisphosphoglycerate phosphatase inhibitor |
| 75 | 0.3 | CYP4A substrate |
| 75 | 0.4 | Hydroxylamine oxidase inhibitor |
| 76.4 | 2.9 | Mucomembranous protector |
| 74.1 | 1.5 | Protein-disulfide reductase (glutathione) inhibitor |
| 73.3 | 1.3 | Fibrinolytic |
| 72 | 0.5 | N-formylmethionyl-peptidase inhibitor |
| 71.5 | 0.4 | Phosphatidylcholine-sterol O-acyltransferase inhibitor |
| 71 | 0.5 | Long-chain-aldehyde dehydrogenase inhibitor |
| 70.6 | 0.4 | Polygalacturonase inhibitor |
Table S4: Biological Activity for n-nonatriacontan-19α-ol
| Percentage Activity | Percentage Inactivity | Name of Activity |
|---|---|---|
| 95.5 | 0.2 | Alkylacetylglycerophosphatase inhibitor |
| 95.3 | 0.2 | Acrocylindropepsin inhibitor |
| 95.3 | 0.2 | Chymosin inhibitor |
| 95.3 | 0.2 | Saccharopepsin inhibitor |
| 94.8 | 0.3 | Sphinganine kinase inhibitor |
| 94.4 | 0.2 | Sugar-phosphatase inhibitor |
| 94.3 | 0.2 | Acylcarnitine hydrolase inhibitor |
| 93.7 | 0.3 | Polyporopepsin inhibitor |
| 93.6 | 0.4 | 5 Hydroxytryptamine release stimulant |
| 93.4 | 0.1 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 93 | 0.1 | Beta-mannosidase inhibitor |
| 92.6 | 0.4 | Ubiquinol-cytochrome-c reductase inhibitor |
| 92.4 | 0.2 | Acetylesterase inhibitor |
| 91.9 | 0.4 | Alkenylglycerophosphocholine hydrolase inhibitor |
| 90.9 | 0.4 | CYP2J substrate |
| 89.3 | 0.3 | Exoribonuclease II inhibitor |
| 88.6 | 0.6 | Benzoate-CoA ligase inhibitor |
| 87.6 | 0.4 | Dextranase inhibitor |
| 88.3 | 1.5 | Membrane integrity agonist |
| 86.6 | 1.3 | Phobic disorders treatment |
| 85.1 | 0.4 | Vasoprotector |
| 85.3 | 0.7 | Prostaglandin-E2 9-reductase inhibitor |
| 84.7 | 0.1 | Sclerosant |
| 84.6 | 0.2 | Leukopoiesis stimulant |
| 84.3 | 0.3 | Alcohol oxidase inhibitor |
| 84.4 | 0.4 | Lipoprotein lipase inhibitor |
| 83.9 | 0.2 | Galactolipase inhibitor |
| 84 | 0.4 | Venombin AB inhibitor |
| 83.7 | 0.3 | N-Acyl-D-aspartate deacylase inhibitor |
| 83.6 | 0.3 | 1,4-Lactonase inhibitor |
| 83.6 | 0.5 | N-acetylneuraminate 7-O(or 9-O)-acetyltransferase inhibitor |
| 83.2 | 0.5 | Ribulose-phosphate 3-epimerase inhibitor |
| 82.4 | 0.4 | Levanase inhibitor |
| 82.1 | 0.4 | Allyl-alcohol dehydrogenase inhibitor |
| 81.7 | 0.2 | Protein-tyrosine sulfotransferase inhibitor |
| 81.5 | 0.3 | Phosphatidylglycerophosphatase inhibitor |
| 80.7 | 0.2 | Oryzin inhibitor |
| 80.1 | 0.4 | Phenol O-methyltransferase inhibitor |
| 80.4 | 0.7 | Omptin inhibitor |
| 80.5 | 0.8 | Arginine 2-monooxygenase inhibitor |
| 79.9 | 0.2 | Platelet aggregation stimulant |
| 80.1 | 0.8 | 5-O-(4-coumaroyl)-D-quinate 3'-monooxygenase inhibitor |
| 79.4 | 0.4 | Lactase inhibitor |
| 79.6 | 0.7 | Superoxide dismutase inhibitor |
| 79 | 0.3 | Aspergillopepsin I inhibitor |
| 78 | 2.3 | Antieczematic |
| 76.4 | 2.9 | Mucomembranous protector |
| 75.4 | 2 | Nicotinic alpha6beta3beta4alpha5 receptor antagonist |
| 73.5 | 0.9 | Lipid metabolism regulator |
| 72.7 | 0.4 | Transketolase inhibitor |
| 72.7 | 0.5 | Oxygen scavenger |
| 73.3 | 1.3 | Fibrinolytic |
| 73.2 | 1.2 | Membrane integrity antagonist |
| 72 | 0.5 | N-formylmethionyl-peptidase inhibitor |
| 71.5 | 0.2 | Antiviral (Rhinovirus) |
| 71.2 | 0.2 | Oxidizing agent |
| 71 | 0.4 | GABA aminotransferase inhibitor |
| 70.5 | 0.8 | Methylumbelliferyl-acetate deacetylase inhibitor |
Table S5: Biological Activity for n-henetetracontan-19α-ol
| Percentage Activity | Percentage Inactivity | Name of Activity |
|---|---|---|
| 95.5 | 0.2 | Alkylacetylglycerophosphatase inhibitor |
| 95.3 | 0.2 | Acrocylindropepsin inhibitor |
| 95.3 | 0.2 | Chymosin inhibitor |
| 95.3 | 0.2 | Saccharopepsin inhibitor |
| 94.8 | 0.3 | Sphinganine kinase inhibitor |
| 94.4 | 0.2 | Sugar-phosphatase inhibitor |
| 94.3 | 0.2 | Acylcarnitine hydrolase inhibitor |
| 93.7 | 0.3 | Polyporopepsin inhibitor |
| 93.6 | 0.4 | 5 Hydroxytryptamine release stimulant |
| 93.4 | 0.1 | Xylan endo-1,3-beta-xylosidase inhibitor |
| 93 | 0.1 | Beta-mannosidase inhibitor |
| 92.6 | 0.4 | Ubiquinol-cytochrome-c reductase inhibitor |
| 92.4 | 0.2 | Acetylesterase inhibitor |
| 92.9 | 0.6 | CYP2C12 substrate |
| 92.1 | 0.1 | Prostaglandin-A1 DELTA-isomerase inhibitor |
| 92.2 | 0.3 | Pro-opiomelanocortin converting enzyme inhibitor |
| 92.1 | 0.3 | Fucosterol-epoxide lyase inhibitor |
| 90 | 0.3 | Cutinase inhibitor |
| 89.3 | 0.3 | Exoribonuclease II inhibitor |
| 88.9 | 0.2 | Gluconate 5-dehydrogenase inhibitor |
| 89.1 | 0.8 | Testosterone 17beta-dehydrogenase (NADP+) inhibitor |
| 88.2 | 0.3 | D-lactaldehyde dehydrogenase inhibitor |
| 88.6 | 0.6 | Benzoate-CoA ligase inhibitor |
| 88.1 | 0.3 | Nitrate reductase (cytochrome) inhibitor |
| 87.6 | 0.4 | Dextranase inhibitor |
| 88.3 | 1.5 | Membrane integrity agonist |
| 87.1 | 0.5 | Feruloyl esterase inhibitor |
| 86.6 | 0.2 | Alcohol dehydrogenase (acceptor) inhibitor |
| 86.9 | 1.5 | Aspulvinonedimethylallyltransferase inhibitor |
| 85.1 | 0.4 | Vasoprotector |
| 85.3 | 0.7 | Prostaglandin-E2 9-reductase inhibitor |
| 84.7 | 0.1 | Sclerosant |
| 84.6 | 0.2 | Leukopoiesis stimulant |
| 84.3 | 0.3 | Alcohol oxidase inhibitor |
| 84 | 0.4 | Venombin AB inhibitor |
| 83.7 | 0.3 | N-Acyl-D-aspartate deacylase inhibitor |
| 83.6 | 0.3 | 1,4-Lactonase inhibitor |
| 82.4 | 0.4 | Levanase inhibitor |
| 81.1 | 0.3 | Snapalysin inhibitor |
| 81.5 | 0.8 | Membrane permeability inhibitor |
| 80.7 | 0.2 | Oryzin inhibitor |
| 80.4 | 0.6 | 2-Hydroxymuconate-semialdehyde hydrolase inhibitor |
| 80.1 | 0.4 | Phenol O-methyltransferase inhibitor |
| 80.4 | 0.7 | Omptin inhibitor |
| 79.4 | 0.4 | Lactase inhibitor |
| 79.6 | 0.7 | Superoxide dismutase inhibitor |
| 79 | 0.3 | Aspergillopepsin I inhibitor |
| 79.1 | 0.5 | Polyamine-transporting ATPase inhibitor |
| 78.4 | 0.2 | Sphinganine-1-phosphate aldolase inhibitor |
| 77.7 | 1.5 | NADPH peroxidase inhibitor |
| 78 | 2.3 | Antieczematic |
| 76.4 | 2.9 | Mucomembranous protector |
| 75.4 | 2 | Nicotinic alpha6beta3beta4alpha5 receptor antagonist |
| 73.3 | 0.6 | Carnitinamidase inhibitor |
| 72.7 | 0.4 | Transketolase inhibitor |
| 72.7 | 0.5 | Oxygen scavenger |
| 73.3 | 1.3 | Fibrinolytic |
| 73.2 | 1.2 | Membrane integrity antagonist |
| 72.2 | 0.4 | GST P substrate |
| 71.5 | 0.2 | Antiviral (Rhinovirus) |
| 71.2 | 0.2 | Oxidizing agent |
| 71 | 0.4 | GABA aminotransferase inhibitor |
| 71.2 | 0.8 | Vasodilator, peripheral |
| 71 | 1.2 | Creatininase inhibitor |
| 71.3 | 1.6 | Glutathione thiolesterase inhibitor |
| 70.1 | 0.5 | D-xylulosereductase inhibitor |
Table S6: Biological Activity for n-heptatriacontan-15α-ol